The antimicrobial peptide database is 20 years old: Recent developments and future directions
Abstract
In 2023, the Antimicrobial Peptide Database (currently available at https://aps.unmc.edu) is 20-years-old. The timeline for the APD expansion in peptide entries, classification methods, search functions, post-translational modifications, binding targets, and mechanisms of action of antimicrobial peptides (AMPs) has been summarized in our previous Protein Science paper. This article highlights new database additions and findings. To facilitate antimicrobial development to combat drug-resistant pathogens, the APD has been re-annotating the data for antibacterial activity (active, inactive, and uncertain), toxicity (hemolytic and nonhemolytic AMPs), and salt tolerance (salt sensitive and insensitive). Comparison of the respective desired and undesired AMP groups produces new knowledge for peptide design. Our unification of AMPs from the six life kingdoms into “natural AMPs” enabled the first comparison with globular or transmembrane proteins. Due to the dominance of amphipathic helical and disulfide-linked peptides, cysteine, glycine, and lysine in natural AMPs are much more abundant than those in globular proteins. To include peptides predicted by machine learning, a new “predicted” group has been created. Remarkably, the averaged amino acid composition of predicted peptides is located between the lower bound of natural AMPs and the upper bound of synthetic peptides. Synthetic peptides in the current APD, with the highest cationic and hydrophobic amino acid percentages, are mostly designed with varying degrees of optimization. Hence, natural AMPs accumulated in the APD over 20 years have laid the foundation for machine learning prediction. We discuss future directions for peptide discovery. It is anticipated that the APD will continue to play a role in research and education.
1 INTRODUCTION
The antimicrobial peptide (AMP) research remains to be active and appealing at least due to the following three reasons: (1) our attempt to decipher the functional roles of AMPs in a variety of organisms; (2) our growing interest in microbiota that can be shaped by AMPs; and (3) our desire of developing AMPs into novel antibiotics (Fjell et al., 2011; Gallo & Hooper, 2012; Hanson et al., 2023; Lazzaro et al., 2020; Salzman & Bevins, 2013). The AMP research may be classified into three main phases. The first phase was started with the discovery of lysozyme in 1922 (Fleming, 1932), which is the first antimicrobial polypeptide according to Robert Lehrer (Personal communication). This early stage of AMP search overlapped with the golden era of antibiotics (1940s–1960s), characterized by the discovery of multiple nonribosomally synthesized peptide antibiotics. Gramicidin was documented as the first peptide antibiotic for clinical use (Dubos, 1939). Colistin, a cyclic lipopeptide, is the last resort of peptide antibiotic against gram-negative pathogenic bacteria (Stansly et al., 1947). Although the discovery of lysozyme did not win Alexander Fleming a Nobel prize, penicillin (discovered in 1928) did. Notably, penicillin is a derivative of the dipeptide cysteine-valine. The second phase was initiated with the discovery of cecropins, defensins, pardaxins, daptomycin, magainins, bactenecin, and histatins in the 1980s (Eliopoulos et al., 1986; Ganz et al., 1985; Gennaro et al., 1989; Oppenheim et al., 1988; Steiner et al., 1981; Thompson et al., 1986; Zasloff, 1987). The invention of two-dimensional nuclear magnetic resonance spectroscopy (2D NMR) and its application in structural determination of small proteins and peptides in the 1970s–1980s (Jeener & Alewaeters, 2016; Wüthrich, 1983) enabled us to see how AMPs look like (Gesell et al., 1997; Wang et al., 2014). The availability of 3D structures of these cationic peptides in membrane-mimetic environments consolidated the amphipathic idea. These structures stimulated the research on the mechanisms of action of antimicrobial peptides, leading to stave-barrel, carpet, detergent, and interfacial models (Ludtke et al., 1996; Oren & Shai, 1998; Vogt & Bechinger, 1999; Wimley, 2010). The research at this phase is also characterized by the efforts in developing magainins into new antibiotics (Jacob & Zasloff, 1994) because it is believed that the rapid membrane disruption by cationic AMPs makes it difficult for pathogens to develop resistance (Zasloff, 2002). The journey in elucidating the molecular mechanism of AMP expression in drosophila resulted in the discovery of Toll and Imd pathways (Lemaitre et al., 1996). This discovery won Jules Hoffmann a Nobel prize in 2011. This milestone discovery inspired the continued search of novel functional roles of these innate immune peptides in coat color, sleep and lifespan till today (Candille et al., 2007; Hanson & Lemaitre, 2023; Toda et al., 2019). The third phase was triggered by the applications of the proteomic and genomic technologies to AMP research. We entered the omic era with the completion of the sequencing of the human genome in 2003 (Human genome project website, visited 2023). The growing research on microbiota added a new dimension to AMP research due to its connection with a variety of human diseases (Wang et al., 2023; Wehkamp et al., 2005). AMPs are important players that shape microbiota (de San et al., 2022; Pierre et al., 2023; Snelders et al., 2021). Remarkably, a Paneth cell peptide YY was able to selectively eliminate pathogenic hyphae but not the commensal yeast form of Candida albicans (Pierre et al., 2023). The omic approaches have also been utilized to AMP discovery (Bishop et al., 2017; Conlon et al., 2006; Lai, 2010; Moyer et al., 2021). Our imagination has gone beyond the classic territories and reached deep in the sea and uncultivable bacteria in soil (Lee et al., 2001; Ling et al., 2015). These studies increased our chances of discovering novel peptide antibiotics to combat drug-resistant bacteria, fungi, viruses, and parasites. The omic approaches have also been applied to the study of immune regulation of host defense peptides, significantly expanding our picture on the roles of AMPs in immune response (Mansour et al., 2014; Mookherjee et al., 2007; Mookherjee et al., 2009).
In the spirit of omics, the Wang laboratory established the antimicrobial peptide database (APD) in 2003 (Wang & Wang, 2004). It is 20-year-old at the time when this manuscript was completed. This database was originally conceived as a tool for antimicrobial and anticancer drug development. Hence, the first version of the APD constructed the following database interfaces: About, Database, Prediction, Peptide design, Statistic Data, Links, and Contact info (to view a picture of the historic website, see Reference Wang, Zietz, et al., 2022). The APD provided the first peptide calculation, empirical prediction, and peptide design tools for AMPs. These database tools were welcomed as echoed by the development of the Grammar approach for peptide design in 2006 and machine learning prediction in 2007, respectively (Lata et al., 2007; Loose et al., 2006). After we demonstrated the template-based design based on our database (Wang et al., 2005), we also conducted database screening to identify novel peptides against human immunodeficiency virus type 1 (HIV-1) or methicillin-resistant Staphylococcus aureus (MRSA; Menousek et al., 2012; Wang et al., 2010). This database provided a statistical tool, allowing users to view the amino acid composition signature for each AMP or a family of peptides (e.g., antibacterial, antiviral, hemolytic, and anticancer). The APD first defined frequently occurring amino acids (>10%), which depend on the source, structure, and activity of AMPs. These abundant amino acids constituted a minimal set for de novo peptide design (Wang et al., 2009). The APD created a powerful search engine by stringing the filters together into a pipeline so that the users can filter the peptide information at their will. Coupled with the statistical information, these database filters laid the foundation for our development of the database filtering technology for designing novel antimicrobials from the beginning (ab initio; Mishra & Wang, 2012a). Our database filtering idea had been extended from in silico to in vitro and in vivo, leading to the discovery of the importance of low cationicity for systemic efficacy (Mishra et al., 2019).
The subsequent two database versions significantly expanded peptide entries, search functions, chemical modification types, 3D structures, binding targets, and mechanisms of action (Wang et al., 2009, 2016). Our database took the lead in systematically annotating over 26 searchable peptide functions (e.g., antibacterial, anti-diabetic, wound healing, anti-toxin, anticancer, antiviral) for two decades. Some of these peptide functions are depicted in Figure 1. In addition, the APD has annotated over 26 types of post-translational modifications of AMPs in a systematic manner (searchable using the XX code; Wang, Zietz, et al., 2022; Wang et al., 2016). A complete list of these functions and chemical modifications can be found in our previous publication (Wang, Zietz, et al., 2022). The APD considered the effects of chemical modifications on peptide properties such as net charge.
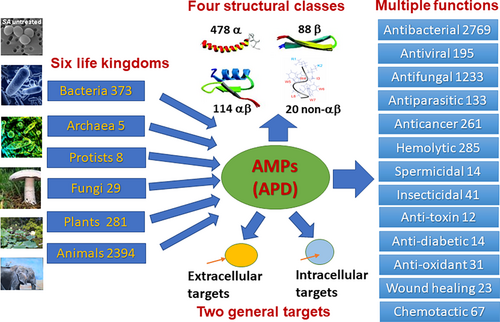
The data scope of the APD evolved with time. While the APD2 collected all peptides reported as AMPs in the literature, including those without activity data (Wang et al., 2009), the APD3 established a set of criteria for data registration by focusing on natural AMPs with known amino acid sequences and activity (minimal inhibitory concentration MIC <100 μM) (Wang et al., 2016). The APD3 also unified the peptide classification methods based on biological sources (six life kingdoms), 3D structures (α, β, αβ, and non-αβ structures), and peptide chain bonding patterns (UCLL, UCSS, UCSB, and UCBB) (Wang, 2022). The six life kingdoms in the APD include bacteria, archaea, protists, fungi, plants, and animals. Currently known AMPs for each kingdom are summarized in Figure 1. The four unified structural classes are α-helical peptides (α class), β-sheet peptides (β class), peptides with both α and β structures (αβ class), and AMPs with neither α nor β structures (non-αβ class). Representative structures for each class are depicted in Figure 1. Due to limited known 3D structures (~13%) for natural AMPs determined by multidimensional NMR spectroscopy and/or x-ray crystallography, the APD also adopted a universal classification scheme based on their covalent bonding patterns. Linear peptides (L) such as human cathelicidin LL-37 and magainins are represented with UCLL (searchable in the name field of the APD). Some linear peptides require two independent chains (LL) to be active. Sidechain–sidechain linked peptides such as defensins (via disulfide bonds) and lantibiotics (via thioether bridges) are searchable using UCSS. A third class (UCSB) has a chemical bond from the sidechain of one amino acid to the C or N-terminus (backbone) of the peptide. Finally, the UCBB peptides contain a chemical bond that connects the N and C-termini of the peptide. In addition, the APD provides a platform for understanding the design principles of natural AMPs (Decker et al., 2022; Lakshmaiah Narayana et al., 2020; Wang, 2020a; Wang, 2020b). Peptides with different activities, structures, and mechanisms of action can all be grouped and analyzed. For instance, the amino acid signatures for AMPs against gram-negative (more lysine) and gram-positive pathogens (more leucine) clearly differ (Wang, 2020a). While leucine is dominant in helical AMPs against gram-positive bacteria, alanine, glycine, and lysine are abundant in helical peptides against gram-negative pathogens (Wang, 2020a). For 1000 amphibian AMPs, there is a linear increase in cationicty (mainly due to lysine) but a decrease in hydrophobic ratio (primarily due to leucine) with peptide length (Wang, 2020b). Remarkably, the averaged arginine content shows a linear correlation with hydrophobic ratio for all the AMPs in the APD (Lakshmaiah Narayana et al., 2020).
Our database expansion, reconfiguration, and information re-annotation continue. This article reports our recent new developments since 2021. These include (1) the unification of the AMPs from the six life kingdoms under the same searchable umbrella “natural AMPs”; (2) the definition of a new “predicted” group for AMPs predicted by machine learning and other methods (e.g., sequence alignment) followed by activity validation; (3) establishment of a systematic scheme for a complete record of reported antimicrobial activity; (4) definition and classification of hemolytic and non-hemolytic AMPs; and (5) definition and systematic annotation of salt effects on peptide activity. Our successful establishment of these new features in the APD not only further enhanced this database capability but also strengthened the notion that our database platform is user-friendly, flexible, and expandable. These re-annotations enabled us to compare natural AMPs with globular proteins and painted the amino acid landscapes for natural, predicted, and synthetic AMPs. Our results shine a novel light on natural AMPs, peptide prediction, and design. Our core data set for natural AMPs accumulated over 20 years has laid the foundation for both prediction and design of novel peptide antibiotics. Finally, we discuss future directions for AMP discovery.
2 NEW FEATURES AND RESULTS
2.1 A look at data growth and peptide parameter space from a new angle
Since the third version (the APD3), natural AMPs have been classified into six life kingdoms: bacteria, archaea, protists, fungi, plants, and animals (current statistics in Figure 1; Wang et al., 2016). The increase of peptide count in each kingdom every 2 years is plotted in Figure 2. In this plot, archaea, protists, and fungi were all located in the valley due to small numbers in the APD (Figure 1). Also, the peptide counts from bacteria and plant kingdoms were below 400. Therefore, the only kingdom that showed a clear increase in peptide number at this scale is animal (72.8% in the APD). Further analysis of the count of animal AMPs every 2 years uncovered a linear correlation (slope 130.88 and R squared 2020: 0.9737). Plant AMPs increased linearly (slope 19.327 and R2: 0.8563) despite at a much slower pace. Likewise, bacterial AMPs grew in proportion to a year over 20 years (slope 22.54 and R2: 0.9788). Hence, natural AMPs, including AMPs from bacteria, plants, and animals, all increased linearly in the APD in the past 20 years. Such linear relationships explain in part why the amino acid signatures for AMPs from bacteria, amphibians, insects, and plants remained the same from 2008 to 2020 (Wang, Zietz, et al., 2022).
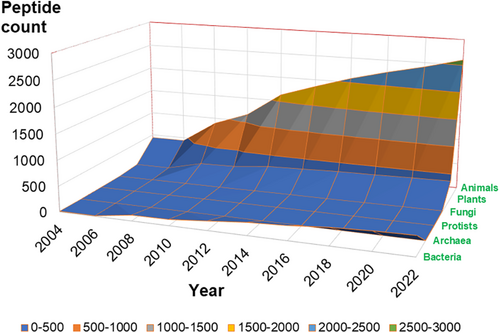
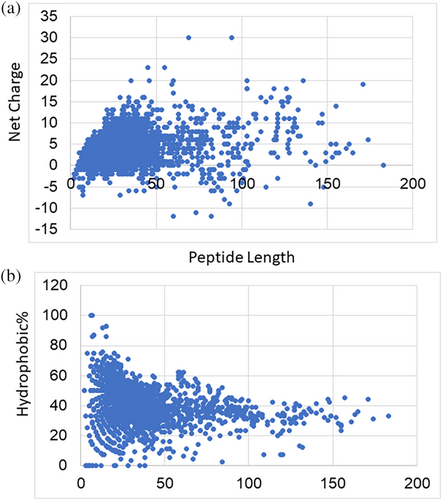
Figure 3 shows the dot plots of net charge or hydrophobic percentage (Pho) as a function of peptide length for all the peptides in the APD. In the case of net charge, the dots were mostly condensed below 50 amino acids and scattered between +30 to −12. Interestingly, short peptides had a low and narrower net charge range (Figure 3A). In contrast, they displayed a wide hydrophobic range. Peptides with a Pho greater than 80% were limited to those with less than 20 residues. The hydrophobic content then shrinked to ~30% with an increase in peptide length (>100 aa; Figure 3B). Examples for AMPs with 0% hydrophobic residues are special peptides. SAAP fraction 3, a surfactant-associated anionic peptide, consists of a string of aspartic acids (Brogden et al., 1996), while shepherins comprised primarily glycine and histidine residues (Park et al., 2000). These are amino acid-rich peptides where at least one amino acid is greater than 25% in the sequence (Decker et al., 2022). Only two cyclic peptides, baceridin, and lugdunin, are entirely hydrophobic (100%; Niggemann et al., 2014; Zipperer et al., 2016). When the peptide length was increased from 0 to 50, there was a clear increase in net charge (Figure 3A) but decrease in hydrophobic percentage (Figure 3B). In the extreme of a low net charge, some peptides such as amphibian temporins (Conlon et al., 2009; Mangoni, 2006) tend to have a high hydrophobic percentage. These hydrophobic peptides, including the two extremes (baceridin and lugdunin), are usually active against gram-positive bacteria such as S. aureus but not gram-negative bacteria such as Escherichia coli.
2.2 Data re-annotation for antimicrobial development
2.2.1 Systematic annotations of antimicrobial activity data
Initially, the APD included very limited activity data by considering the challenging nature in comparing the activity of AMPs conducted in different laboratories using different methods and conditions. So, AMPs were qualitatively labeled as antimicrobial (antibacterial, antiviral, antifungal, and anticancer) in the first version of the APD (Wang & Wang, 2004). With the increased research on microbiota that requires selective peptide antibiotics, it became necessary to more completely annotate the activity of AMPs. This is because microbiota is so elegant that even one invading pathogen can tilt its balance. Hence, a peptide in principle can be tailored to correct the imbalance if it can perform a targeted elimination (Kim et al., 2019; Li et al., 2021). In 2021, the APD created a complete activity annotation system based on our previous definition for active and inactive peptides (Wang, Zietz, et al., 2022). This annotation system enables a more complete record of the reported activity data for each AMP. In this system, peptide activities were classified into three categories: active peptides (MIC <100 μM), inactive peptides (MIC > 100 μM), and uncertain (activity greater than an MIC value less than 100 μM) (summarized in Table 1). Active AMPs were annotated in the “Additional Information” using the abbreviated form of scientific names (e.g., E. coli, S. aureus, and C. albicans). When E. coli was entered into “Additional Information” followed by “search, we obtained 1834 AMPs active against E. coli. In contrast, peptides that did not kill a specific microbe (MIC > 100 μM) are annotated in a different format so that this set of inactive peptides could be searched (e.g., E.coli, S.aureus, and C.albicans, no space). For example, we obtained 218 peptides when we searched the database using E.coli (Table 1). Those AMPs with uncertain activity (e.g., 100 μM > MIC > any value) were annotated in a third format (e.g., E-coli, S-aureus, and C-albicans). In this way, all the three types of peptide activities could be searched in the APD via the “Additional information” field. Table 1 tabulated the three types of peptide activities annotated to date against E. coli, S. aureus, and C. albicans. Any other microbes such as P. aeruginosa can be searched in the same manner as long as its activity has been entered into the APD.
| Activity type | Active | Inactive | Uncertain |
|---|---|---|---|
| Definition | MIC < 100 μM | MIC > 100 μM | For example, MIC > 16 μM but not > 100 μM |
| Microbe annotation and search formata | E. coli, or S. aureus | E.coli, or S.aureus | E-coli, or S-aureus |
| Escherichia coli | 1834 | 218 | 62 |
| Staphylococcus aureus | 1744 | 161 | 88 |
| Candida albicans | 806 | 163 | 31 |
- a Data obtained from the “Additional information” field of the database (https://aps.unmc.edu) as of July 2023 using the defined search format. Although only three commonly used microbes were utilized here to illustrate the status of data annotations, any microbe species can be searched in the same manner.
There are more data for E. coli, S. aureus, and C. albicans in the APD due to wide use of these strains in antimicrobial screening. Hence, we first compared the amino acid signature of active and inactive AMPs against E. coli. In the case of helical peptides, those active against E. coli had clearly higher contents of lysine and arginine, although glycine and valine were only slightly higher. In contrast, those helical AMPs with very weak or no activity against E. coli possessed higher contents of alanine, phenylalanine, isoleucine, leucine, and serine (summarized in Table 2). Clearly, cationic amino acids play an important role in killing E. coli. We then compared AMPs active and inactive against S. aureus. There appeared an opposite requirement for S. aureus. Active helical AMPs showed higher contents of phenylalanine, glycine, and leucine, while those S. aureus-inactive helical AMPs were higher in lysine and alanine (Table 2). These amino acids were derived from the amino acid plots (Figure S1). These plots support the contention that hydrophobic amino acids are critical in inhibiting gram-positive MRSA. This database finding is consistent with the observation from structure–activity relationship (SAR) studies (Li et al., 2006; Wang et al., 2018). Interestingly, helical AMPs active against C. albicans resembled those against E. coli since they also had higher contents of lysine and arginine. Such a feature differed from those C. albicans-inactive peptides with higher percentages of hydrophobic leucine and alanine.
| Group | Desired | Undesired | Data source | |
|---|---|---|---|---|
| 1 | Peptide target | Antibacterial | Non-antibacterial | |
| E. coli | K, R, V | A, F, I, L, S | Figure S1A | |
| S. aureus | F, G, L, P | A, K, Y | Figure S1B | |
| 2 | Peptide scaffold | Non-hemolytic | Hemolytic | |
| UCLL | V, W, (P), Q, (N), K, R | I, L, (F), A, G, S, (H) | Figure S2A | |
| UCSS | M, (W), G, N, D, H, R | I, (V), (L), F, C, (A), P, T, K | Figure S2B | |
| 3 | Peptide scaffold | Salt-insensitive | Salt-sensitive | |
| UCLL | F, G, P, H, R | A, S, Q, N, E, D, K | Figure S3A | |
| UCSS | V, L, T, S, N, H, R | I, A, P, Q, E, D, K | Figure S3B |
- a Significantly different in bold, and smaller changes in parenthesis. Data obtained from the APD (https://aps.unmc.edu) in July 2023. Figure S can be found in the Supporting Information section of this article.
We then compared active and inactive AMPs with beta structures. A consensus picture emerged irrespective of the pathogen type: E. coli, S. aureus, or C. albicans. Such a similarity for β-sheet AMPs may be attributed to the broad-spectrum activity of defensin-like peptides. The active groups with a beta structure were abundant in cysteine, glycine, and arginine, three critical amino acids in defensins (10.5% R for 379 defensins). However, the three dominant amino acids in the inactive groups were cysteine, glycine, and threonine (Table 2). The low content of arginine (4.3% or less) might be one of the major reasons for the lack of activity of the inactive group since cysteine and glycine are the two common amino acids required for β-sheet structure formation.
2.2.2 Hemolytic and nonhemolytic peptides
The development of AMPs into a new generation of antibiotics requires that the peptide do a targeted pathogen elimination without harming the host (Bobde et al., 2021; Hancock et al., 2021). Hemolysis is frequently utilized to gauge peptide cytotoxicity. Hemolytic peptides were annotated in the first version of the APD (Wang & Wang, 2004). Our statistical analysis at that time revealed a higher hydrophobic content for hemolytic peptides than other antimicrobial groups. To better understand the differences between hemolytic and nonhemolytic AMPs at the amino acid level, the APD is in the process of re-annotating the peptides by defining criteria for “hemolytic” and “non-hemolytic” as well. Hemolytic peptides show a clear dose-dependent hemolysis with a 50% hemolytic concentration (HC50) less than 100 μM, while nonhemolytic peptides show no sign of hemolysis at least at 10-fold of MIC and have a HC50 value greater than 100 μM. In many cases, however, a detailed hemolytic curve is not provided in published papers and only HC50 may be available for this estimation. Like antimicrobial activity, the APD is fully aware of the challenge in the heterogeneity of hemolytic data, which can be influenced by numerous factors, including protocols, blood cell types (e.g., human and nonhuman), container (Eppendorf tubes or microplate/type), blood and peptide freshness, cell concentration, and incubation time. Preferred amino acids for hemolytic and nonhemolytic AMPs currently annotated in the APD are provided in Table 2 as well. These amino acids were derived from the amino acid plots of these two groups (Figure S2). It is evident that hemolytic peptides from the linear class (UCLL) possessed higher contents of hydrophobic amino acids, including isoleucine, leucine, phenylalanine, and alanine. In contrast, the non-hemolytic group showed higher contents of lysine, arginine, tryptophan, valine, glutamine, and asparagine. We also compared the two sidechain-linked AMP (UCSS) groups with hemolytic and non-hemolytic activities. Again, there were more hydrophobic residues I and F in the hemolytic group than the those in the nonhemolytic group. The nonhemolytic group had a substantial percentage of glycine, asparagine, and arginine. It appeared that arginine was higher in both nonhemolytic groups irrespective of the structural scaffolds. These comparisons may inspire the design of nonhemolytic peptides.
2.2.3 Consensus amino acids in salt-sensitive and insensitive antimicrobial peptides
It is known that some AMPs could lose antimicrobial activity in the presence of physiological salts (Huang et al., 2007; Krishnakumari et al., 2013). There are some studies in the literature aiming at designing AMPs resistant to salts (Chu et al., 2020; Dou et al., 2017; Mishra et al., 2019). However, a global picture for salt resistance of AMPs was lacking. The APD started to pay attention to this information before 2018, especially after the successful extension of our in silico filters to in vitro filters (Mishra et al., 2019). The in vitro filters include salt tolerance, pH sensitivity, and serum binding, which are known hurdles that might cause a drop or even loss of peptide activity. While data are accumulating, we documented herein the amino acid differences between the salt-sensitive and insensitive groups of AMPs currently annotated in the APD. Salt-sensitive AMPs changed their MIC value four-fold or more in the presence of 150 mM NaCl, while the MIC values of salt-insensitive peptides remained constant (not more than two-fold change in MIC). We narrowed down our database search to AMPs less than 50 amino acids and further split them into two structural classes: UCLL and UCSS based on our universal peptide classification (Wang, 2022). The results are included in Table 2. The UCLL class consisted of linear AMPs (22 salt-insensitive vs. 9 salt-sensitive), which might form amphipathic helices upon interaction with bacterial membranes. Typical examples in the UCSS class are defensins (20 salt-sensitive vs. 6 salt-insensitive). In the case of the linear group, both arginine and histidine were higher in the salt-resistant group (i.e., salt-insensitive in Table 2) than in the salt-sensitive group. Likewise, these two amino acids were also more abundant in the salt-resistant sidechain-linked group than the salt-sensitive group. In addition, the salt-sensitive AMPs shared a consensus set of abundant amino acids (A, Q, E, D, and K). These amino acids were derived from the amino acid signature plots for the respective groups (Figure S3). It appears that there was a consensus in arginine preference irrespective of the peptide structural class. Such a discovery may inspire the design of salt-resistant AMPs.
2.3 New light on natural antimicrobial peptides by comparison with globular proteins and transmembrane proteins
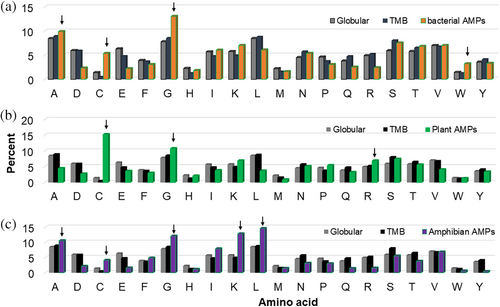
The first version of the APD enabled statistical analysis of AMPs. When the percentages of the 20 amino acids with different antimicrobial activities were plotted, it became clear that some amino acids (e.g., cationic K and R) are more abundant than other amino acids (e.g., acidic D and E). This explains why most of the AMPs in the APD are cationic (94.4% with net charge ≥0). However, we have never compared the amino acid composition of natural AMPs with that of globular or transmembrane proteins. To facilitate this comparison, we made a new addition to the APD by manually annotating relevant peptide entries with a searchable indicator “natural AMPs,” which includes all peptides from the six life kingdoms (Figure 1; Wang et al., 2016). This led to a total of 3090 natural AMPs when we conducted this search in the name field using “natural AMPs.” This list did not include 208 natural peptides without activity data (annotated as DXWZ), which were collected into the APD prior to the definition of data registration criteria. We then compared the amino acid compositions of natural AMPs with globular and transmembrane proteins (not shown; Gromiha et al., 2005). Lysine, glycine, and cysteine of natural AMPs on average were higher than those in either globular or transmembrane proteins (which are similar). These three residues resulted from a combination of dominant amino acids in α-helical (K and G) and β-sheet AMPs (G and C) in this database (Mishra & Wang, 2012a; Wang & Wang, 2004). To gain additional understanding, we also compared the amino acid composition of globular proteins with those of AMPs from bacteria, plants, and amphibians. In the case of bacterial AMPs (Figure 4A), cysteine and glycine were clearly higher, while leucine, arginine, aspartic, and glutamic acids were much lower. Cysteine in plant AMPs was extremely high due to the fact that these peptides frequently form disulfide bonds (Figure 4B). In addition, lysine, arginine, and glycine were slightly higher while aspartic and glutamic acids were lower. In the case of amphibian AMPs, residues alanine, glycine, leucine, and lysine became dominant (Figure 4C). These are the known frequently occurring amino acids we initially discovered in the APD2 paper (Wang et al., 2009). Beyond these amino acids, threonine was also clearly lower, but isoleucine was higher. In addition, tyrosine, arginine, glutamine, glutamic, and aspartic acids were extremely low compared to those in globular proteins. It appeared that these amino acids were less preferred in nature's design of amphibian AMPs. Thus, those more abundant amino acid signatures differed: A, C, G, and W for bacterial AMPs, C, G, and R for plant AMPs, and A, C, G, K, and L for amphibians (Figure 4 arrows). While C and G were higher in all the three AMP groups than that in globular proteins, D and E were consistently lower. Also, the use of tryptophan differed: more in AMPs from bacteria than from amphibian. Our new results obtained herein provided additional insight into AMP design in different life kingdoms or classes since amino acid compositions play an important role in determining peptide structure and activity spectrum (Maasch et al., 2023; Mishra & Wang, 2012a; Mishra & Wang, 2012b; Wang, Vaisman, & van Hoek, 2022).
2.4 A new peptide group is created in the APD for machine learning-predicted antimicrobial peptides: Amino acid landscapes of natural, predicted, and synthetic AMPs
Since the first single-label prediction of AMPs by machine learning in 2007 (Lata et al., 2007), the APD has also been utilized for multi-label prediction (Xiao et al., 2013) based on a variety of peptide functions/activities annotated therein (Figure 1). Machine learning prediction is becoming popular these days (Maasch et al., 2023; Wang, Vaisman, & van Hoek, 2022; Xiao et al., 2013). It opens a new avenue for AMP discovery. Even encrypted antimicrobial peptides from extinct paleoproteomes were predicted by this technology in 2023 (Maasch et al., 2023). Hence, the APD cannot ignore new peptides from these predictions, especially those predicted from genomes. As we do not have enough knowledge to judge to what extent these predicted sequences correspond to natural AMPs, we decided to create a new group “predicted” for these peptides if their antimicrobial activity had been proved experimentally (i.e., MIC <100 μM). Note that the predicted AMPs in the APD also include those sequences predicted by other methods, such as sequence alignment. For instance, FALL-39, initially included in the LL-37 entry (AP00310), now occupies an independent entry (AP03566) as a classic member of the predicted family based on the highly conserved precursor sequences of cathelicidins (Agerberth et al., 1995). FALL-39 differs from LL-37 by two residues and ALL-38 by only one residue. Both LL-37 (other tissues such as blood neutrophils and skin) and ALL-38 (from human reproductive system) are isolated AMPs (Gudmundsson et al., 1996; Sørensen et al., 2003). Likewise, one of the two predicted forms of murine cathelicidin differs from the isolated one by only one residue (Pestonjamasp et al., 2001). These results indicate that predictions based on sequence alignment can reach a remarkably high accuracy for cathelicidins (95%–97%). At the moment, we do not have data for a similar accuracy comparison of machine learning-predicted peptides with natural AMPs. By the time this manuscript was completed, the APD has registered 194 peptides predicted by different methods, mostly AI predicted.
We then asked how the predicted peptides were deviated from natural AMPs. Figure 5 compares the averaged amino acid composition profile of natural AMPs with that of predicted peptides in the APD. The overall trend was remarkably similar, indicating heavy dependence of the positive training data of natural AMPs we obtained in the past 20 years. However, residues K and R were much more abundant in predicted AMPs (blue), while residues D and E were lower. Meanwhile, hydrophobic residues F, I, L, and V were higher, but A and C were lower than those in natural AMPs. Such a combined tilt toward larger hydrophobic and basic amino acids by machine learning might have increased the success of predicted AMPs. A lower C implied an emphasis of the current predictions on linear sequences.
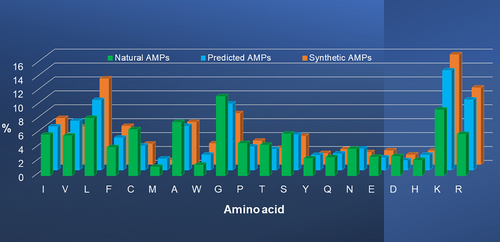
Synthetic peptides are human-made AMPs to understand structure–activity relationships or to improve antimicrobial therapeutic potential. They form a separate group in the APD from the birth of the database in 2003. Due to our previous focus on natural AMPs, not all synthetic peptides have been collected yet. Among the 121 synthetic AMPs in the APD, many were designed using amino acids known to be important for peptide activity. In addition, some synthetic AMPs have been optimized for high activity and low toxicity. Library screen and SAR studies are two common in vitro evolution methods to make the candidate peptide more potent and less toxic (Blondelle et al., 1995; Deslouches et al., 2005; Jacob & Zasloff, 1994; Lakshmaiah Narayana et al., 2020; Loose et al., 2006; Mishra et al., 2019). For instance, combi-1 and combi-2 are among the best peptides discovered from combinatory libraries (Blondelle et al., 1995), while WLBU2 is obtained from a de novo design based on valine and arginine (Deslouches et al., 2005). Likewise, the two best peptides D28 and D51 from the grammar approach are found in the APD (Loose et al., 2006). Horine and verine are optimal candidates identified from a family of database-designed peptides (Lakshmaiah Narayana et al., 2020). We were curious how the plot for these synthetic peptides looked like. Remarkably, residues I, L, F, W, K, and R were highest, while residues A, G, N, S, and T were lowest (gold in Figure 5) compared to both predicted and natural AMPs. Hence, a select set of basic and hydrophobic amino acids was elevated to the highest level in the synthetic peptides in the current APD. Again, residue C was lowest in synthetic peptides, implying a preference of linear peptides. M was lowest to avoid loss of activity from auto-oxidation.
It is interesting that the amino acid composition of the predicted AMPs on average lay between those of natural and synthetic AMPs (Figure 5). Since both natural and synthetic peptides in the APD had usually been included in machine learning predictions, they might have set the lower and upper parameter bounds for such predictions. The picture is clear that natural AMPs laid the foundation for both synthetic and predicted peptides, which had higher cationic (K and R) and hydrophobic amino acids (I, L, F, and W) for potency. This was achieved by decreasing small amino acids (A, G, and S). W is low in natural AMPs (Figure 5) but has been widely incorporated into synthetic AMPs, especially short ones (Blondelle et al., 1995; Chan et al., 2006; Cherkasov et al., 2009; Lakshmaiah Narayana et al., 2020). We conclude that the classic amphipathic concept is deeply rooted in the prediction and design of AMPs.
3 CONCLUSION AND FUTURE DIRECTIONS
The APD is a founding database for the AMP field first constructed based on the LAMP software bundle, including Red Hat Linux operating system using the freeware Apache web server, MySQL relational database management system, and PHP script language (Wang & Wang, 2004). It built numerous database features for the first time, including unified classifications of AMPs in numerous ways (Figure 1) (Wang, Zietz, et al., 2022). Consequently, the APD is widely utilized in research and education (Wang et al., 2016). To facilitate education, the APD has put information in order by establishing multiple educational web pages, including peptide discovery timeline, glossary, nomenclature, classification, structural determination, tools, and web links. As an educational event, a hybrid symposium entitled “Antimicrobial Peptides: Yesterday, Today and Tomorrow” will be held in Omaha, Nebraska, USA on October 6, 2023.
To further advance AMP research, this study has unified the AMPs from bacteria, archaea, protists, fungi, plants, and animals under a common umbrella “natural AMPs” (searchable). Our new annotation facilitated a comparison of natural AMPs with globular proteins, providing novel insight into the amino acid use in natural AMPs in general and in bacteria, plants, and amphibian AMPs in particular. Different from globular proteins (usually > 100 amino acids), which automatically fold under physiological conditions, many AMPs (usually < 50 amino acids) are forced to a structural scaffold via disulfide bond formation. A majority of linear AMPs are disordered in aqueous solution and only fold into an amphipathic helical structure upon binding to membranes. The amphipathic concept has been dominating the field and widely utilized as a general principle to design and improve AMPs for decades. However, some linear peptides (e.g., proline-rich AMPs) may also adopt a non-helical structure after association with ribosomes (Gagnon et al., 2016).
This study also expanded the scope of AMPs in the APD by creating a new group predicted by machine learning followed by antimicrobial validation. However, peptides predicted by other methods such as sequence alignment are also included in this group. The similarity in amino acid landscapes (Figure 5) underscores that the natural AMPs accumulated in the APD over 20 years have laid the foundation for machine learning prediction. It is interesting to note that, man-made peptides, be they predicted or designed, are deviated from the baseline set by natural AMPs due to higher cationic and hydrophobic amino acid incorporation to enhance antimicrobial potency. Synthetic peptides in the APD, despite a small set, have been designed and optimized to varying degrees, thereby setting the parameter upper bound for peptide prediction.
Our data re-annotation has generated new knowledge for AMPs. Cationic amino acids are important for antimicrobial activity of helical peptides against gram-negative E. coli, while leucine, phenylalanine, and glycine are more abundant in helical peptides active against gram-positive S. aureus, including MRSA. This is in line with our previous discovery that high hydrophobicity is important for anti-MRSA peptides (Mishra et al., 2019). Along this line, there are extremes where some anti-MRSA AMPs are entirely hydrophobic, indicating charged amino acids are not a must for killing MRSA. These hydrophobic extremes, together with hydrophilic extremes (e.g., poly-aspartic AMPs), provide exceptions to the amphipathic concept. Arginine plays a critical role in β-sheet AMPs broadly active against E. coli, S. aureus, and C. albicans. Among the multiple arginines in a central peptide of human LL-37, only the sidechain of R23 can more effectively interact with anionic lipid headgroup (Wang, 2007). Arginine is important for antiviral activity as well (Wang et al., 2010; Wang & Wang, 2004). This study suggests that arginine might be useful in conferring salt resistance to AMPs (Table 2). In certain cases, arginine may also make the peptide more hemolytic to human erythrocytes as it is more hydrophobic than lysine (Mishra, Lushnikova, et al., 2017). While the first version of the APD uncovered higher hydrophobic contents for hemolytic AMPs (Wang & Wang, 2004), our data re-annotation here reveals that high hydrophobicity for hemolytic peptides results from amino acids F, I, and L. Hence, this study has enriched our knowledge on the design of novel AMPs.
Before closing, it is also useful to point out some possible directions for AMP discovery. In the APD3 paper, we projected the number of natural AMPs at least in the million range if each of the 1.3 million named species can produce at least one such peptide (Wang et al., 2016). After re-annotation, the APD contained 3090 “natural AMPs” (with activity data) out of a total of 3623 peptides as of April 2023. Therefore, the AMP field has a long way to go to reach 1 million natural AMPs. Future studies may explore new AMPs in those under-represented life kingdoms, including bacteria, archaea, fungi, protists, and plants (Figure 2 valley). Indeed, scientists are discovering new AMPs from bacteria in a variety of environments, be they deep in the sea, buried in soil, or hidden in animal guts (Hansen et al., 2020; Hatziioanou et al., 2017; Ling et al., 2015; Ma et al., 2022). We anticipate the application of various methods ranging from the classic isolation and purification to machine learning in future peptide discovery. With the discovery of additional AMP members from those under-represented kingdoms and further development of artificial intelligence algorithms, we anticipate a great advance in peptide prediction and design toward future antibiotics, food preservatives, and “green” pesticides. To restore imbalanced microbiota, species-specific AMPs are of top interest. This is a challenging task since it requires the use of many more relevant bacterial strains in antimicrobial assays to define the activity spectrum of a particular candidate before use. Our new data annotation scheme (Table 1) has reached a more complete record of antimicrobial activity data. This will facilitate activity spectrum matching for potential therapeutics. Based on the current knowledge, bacteriocins (i.e., bacterial AMPs) constitute excellent candidates to correct the imbalance of microbiota because of their frequently observed species-specific antimicrobial activity. Therefore, we anticipate that future AMP research will pay more attention to bacteriocins with a variety of structural scaffolds. Indeed, several AMPs currently in clinic use or for food preservation are made by microbes (Mishra, Reiling, et al., 2017).
In summary, the APD has been evolving for 20 years with continued updates and refinement. This study has further expanded the landscape of AMPs registered in the APD and improved its capability for antimicrobial development. With further developments in the future, the APD will continue to serve the antimicrobial research community.
AUTHOR CONTRIBUTIONS
Guangshun Wang: Conceptualization (equal); data curation (equal); formal analysis (equal); funding acquisition (equal); project administration (equal); supervision (equal); validation (equal); writing – original draft (equal).
ACKNOWLEDGMENTS
The author appreciates previous grants as well as the current National Institutes of Health grants GM138552 and AI175209. The author is grateful to the department of Pathology and Microbiology as well as UNMC IT for years of support. I thank my first graduate student Zhe Wang who did a wonderful job in creating the original APD. We appreciate all authors for peptide deposition and all users for additions, corrections, and suggestions.
CONFLICT OF INTEREST STATEMENT
The author declares no conflicts of interest.




