Phosphorylation of Schizosaccharomyces pombe Dss1 mediates direct binding to the ubiquitin-ligase Dma1 in vitro
Abstract
Intrinsically disordered proteins (IDPs) are often multifunctional and frequently posttranslationally modified. Deleted in split hand/split foot 1 (Dss1—Sem1 in budding yeast) is a highly multifunctional IDP associated with a range of protein complexes. However, it remains unknown if the different functions relate to different modified states. In this work, we show that Schizosaccharomyces pombe Dss1 is a substrate for casein kinase 2 in vitro, and we identify three phosphorylated threonines in its linker region separating two known disordered ubiquitin-binding motifs. Phosphorylations of the threonines had no effect on ubiquitin-binding but caused a slight destabilization of the C-terminal α-helix and mediated a direct interaction with the forkhead-associated (FHA) domain of the RING-FHA E3-ubiquitin ligase defective in mitosis 1 (Dma1). The phosphorylation sites are not conserved and are absent in human Dss1. Sequence analyses revealed that the Txx(E/D) motif, which is important for phosphorylation and Dma1 binding, is not linked to certain branches of the evolutionary tree. Instead, we find that the motif appears randomly, supporting the mechanism of ex nihilo evolution of novel motifs. In support of this, other threonine-based motifs, although frequent, are nonconserved in the linker, pointing to additional functions connected to this region. We suggest that Dss1 acts as an adaptor protein that docks to Dma1 via the phosphorylated FHA-binding motifs, while the C-terminal α-helix is free to bind mitotic septins, thereby stabilizing the complex. The presence of Txx(D/E) motifs in the disordered regions of certain septin subunits may be of further relevance to the formation and stabilization of these complexes.
1 INTRODUCTION
Deleted in split hand/split foot 1 (Dss1—Sem1 in budding yeast) is a small, eukaryotic, conserved, and intrinsically disordered protein (IDP) that serves as a subunit of the large 26S proteasome complex (Funakoshi et al., 2004), where it is important for its assembly (Jossé et al., 2006; Kolog Gulko et al., 2018; Tomko & Hochstrasser, 2014) and allosteric regulation of substrate unfolding (Reed et al., 2022). Yeast mutants lacking Dss1 display a temperature-dependent growth defect largely linked to faulty proteasome assembly (Tomko & Hochstrasser, 2014). Dss1 is multifunctional and has been found to engage in a broad interactome with functions in DNA repair (Yang et al., 2002), mRNA export (Faza et al., 2009), and other cellular pathways (Schenstrøm et al., 2018), and has also been reported to regulate the assembly of other protein complexes (Le et al., 2020; Lee et al., 2021; Pick et al., 2009). In humans, Dss1 has been suggested to be implicated in different types of cancers including breast, colorectal, and cervical cancers (Achille et al., 1996; Ma et al., 2013; Rezano et al., 2013). In the fission yeast Schizosaccharomyces pombe (S. pombe) Dss1 has been identified as an IDP, capable of binding ubiquitin (Paraskevopoulos et al., 2014) via two disordered ubiquitin-binding motifs covering the regions D16-N25 (DisUBM2) and D38-D49 (DisUBM1) (Dreier et al., 2022), an ability conserved in human Dss1 (Ruidiaz et al., 2021). The two ubiquitin-binding sites are positioned in two out of three conserved sequence regions in the Dss1 protein, separated by a non-conserved linker (Figure 1). The ability of Dss1 to bind ubiquitin may play a role in several of the biological functions of Dss1, constituting functional links between different multiprotein complexes, in which it acts as a scaffold protein to stabilize their architectures (Kragelund et al., 2016).
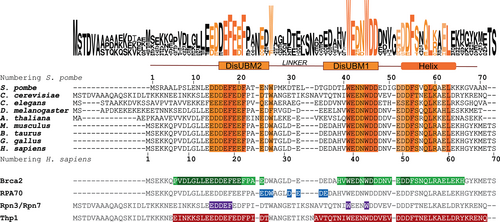
A third conserved region of S. pombe Dss1 is located in its C-terminal region, where it forms a transiently populated α-helix from F55 to K66 (Paraskevopoulos et al., 2014). This helix has been shown to dynamically contact the non-conserved linker region in a fold-back orientation through which it can restrict access to DisUBM1 (Paraskevopoulos et al., 2014). As a consequence, removal of the helix causes a rewiring of the interactome of Dss1, as identified by proteomics (Schenstrøm et al., 2018). Besides the approximately 50% populated α-helix, Dss1 is otherwise devoid of secondary structure and because of its high negative net charge of −18 at physiological pH, it adopts an extended and highly dynamic structure in its unbound state (Newcombe et al., 2021; Pesce et al., 2023). The α-helix and the two conserved regions harboring the DisUBMs have been found to interact with many binding partners (Schenstrøm et al., 2018; Figure 1), including calcium ions (Newcombe et al., 2021). The three regions are the main interaction sites in Dss1, while the non-conserved linker region is the primary contact point for replication protein A (RPA; Liu et al., 2010; Zhao et al., 2015; Figure 1). Thus, despite the poor conservation of the linker sequence stretch, it is possible that other Dss1 ligands may engage with this region. Indeed, the linker region varies highly in length and composition across species. As it connects the two DisUBMs, the linker length may impact binding via the DisUBMs and may confer specificity to specific types of ubiquitin-chain linkages.
Phosphorylation is one of the most common posttranslational modifications (PTMs; Iakoucheva et al., 2004; Khoury et al., 2011; Tyanova et al., 2013). It is capable of inducing structural changes both locally and globally and particularly in IDPs (Bah et al., 2015; Hendus-Altenburger et al., 2017). Through a number of different mechanisms including structural modulation phosphorylation may alter protein interactions (Newcombe et al., 2022). As an IDP, Dss1 is likely to be regulated by PTMs, but no phosphorylation sites or other PTMs, including ubiquitinylation of Dss1, have been reported. Notably, the sequence of Dss1 contains several serines and threonines, which may serve as potential phosphorylation sites. However, these are not evolutionarily conserved and their relevance to the function of Dss1 is unclear.
In this study, we show that S. pombe Dss1 is a substrate for casein kinase 2 (CK2) in vitro and identify three threonines in its linker region as phosphorylation targets. Phosphorylation of these residues did not affect mono-ubiquitin binding but destabilized the C-terminal α-helix slightly. However, phosphorylation was found to mediate direct interaction between Dss1 and the forkhead-associated (FHA) domain of the RING E3-ubiquitin ligase defective in mitosis 1 (Dma1), previously identified as a Dss1 interaction partner by proteomics (Schenstrøm et al., 2018). Sequence analysis showed that all three phosphorylation sites within the linker of Dss1 have the short linear phosphorylation motif Txx(E/D). The three motifs are not conserved across species and have likely arisen ex nihilo. We suggest that Dss1 may act as an adaptor protein for Dma1, where it uses its phosphorylated FHA-binding motifs in the linker to dock to Dma1, while its C-terminal α-helix may bind to the septins, that in some species are substrates of Dma1 (Chahwan et al., 2013). This property of Dss1 appears to be lost in human and budding yeast orthologues. We also make the observation that the disordered regions in septins similarly carry the Txx(E/D) motif suggesting diverse mechanisms for recruitment of Dma1.
2 RESULTS
2.1 Dss1 is a substrate for casein kinase 2
We first analyzed the sequence of Dss1 for putative phosphorylation sites. Different serines and threonines were suggested as substrates by different predictors. Kinase-specific prediction tools (NetPhosK, GPS3.0, and Scansite 3; Blom et al., 1999; Obenauer et al., 2003; Wang et al., 2020) predicted phosphorylation of T35 by casein kinase 2 (CK2), while three out of five tools predicted phosphorylation of T39 and T31, two of these with CK2 as the kinase. Thus, the most likely phosphorylation sites of Dss1 appeared to be T35, T39, and T31 (in order of decreasing prediction score) with the most likely kinase being CK2. CK2 is a ubiquitous, abundant, and conserved protein kinase with substrates in both the cytosol and the nucleus (Faust et al., 1999), and it can act as a primed kinase in the presence of pThr and pSer (St-Denis et al., 2015). It phosphorylates proteins involved in the regulation of DNA repair (Litchfield, 2003), gene expression, signaling, apoptosis and many other cellular processes (Selenko et al., 2008). Unlike many kinases, CK2 is constitutively active and independent of prior phosphorylation events or second messengers (Meggio & Pinna, 2003). The analysis of >300 confirmed phosphorylation sites established a minimum consensus sequence for CK2: [S/T]xx[E/D], with a preference for serine over threonine (Meggio & Pinna, 2003). Acidic residues at positions n + 3 and n + 1 (with n being the phosphorylated residue) occur in 90% and 75% of the phosphorylation sites, respectively. Phosphorylation sites lacking an acidic residue at one position always have it at the other position (Meggio & Pinna, 2003). The three predicted sites in Dss1 adhere to these preferences with an acidic residue at the n + 3 position, and with T31 having one also at n + 1 (Figure 2a).
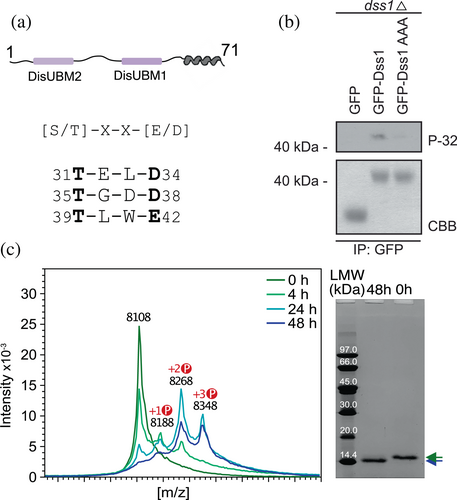
We next asked if Dss1 is phosphorylated in cells. Wild-type (WT) Dss1 was overexpressed as a GFP fusion protein from a plasmid in S. pombe cells deleted for endogenous dss1 (dss1Δ). Radiolabeling with 32P orthophosphate followed by immunoprecipitation using GFP-trap resin showed that Dss1 was phosphorylated (Figure 2b). As a control, a Dss1AAA variant was tested, where the three consensus threonines were replaced by alanines. Here, phosphorylation was strongly reduced, confirming the phosphorylation sites to be one or several of the three Thr residues. As an additional control for the presence and purity of the proteins, GFP, GFP-Dss1 WT, and the GFP-Dss1AAA variant were purified from larger (unlabeled) cultures, and analyzed by SDS-PAGE (Figure 2b, bottom). We also attempted to address if CK2 would be the kinase responsible for this phosphorylation in vivo, and used the cka1 mutant (orb5-19) to do so. However, in our hands, this mutant displayed a poor growth, and we did therefore not pursue this further. Instead, we tested if CK2 would phosphorylate Dss1 in vitro. After incubation of recombinantly purified Dss1 with active CK2 in the presence of ATP, we analyzed the sample with both SDS-PAGE and MALDI-TOF mass spectrometry (MS). For the SDS-PAGE, we observed an electrophoretic mobility shift, consistent with the notion that Dss1 becomes increasingly negatively charged upon its phosphorylation, binding less SDS and migrating slower in the gel, as observed for other systems previously (Lee et al., 2019; Figure 2c). When we analyzed the samples with MALDI-TOF MS, we detected three peaks with mass shifts of 80 Da increments appearing in the spectra compared to the t0 control, which are consistent with phosphorylation events (Figure 2c). Taken together, these results highlight that Dss1 is likely a physiological substrate of CK2, carrying up to three phosphorylation sites.
2.2 Thr31, Thr35, and Thr39 are phosphorylated by CK2 in a distinct order
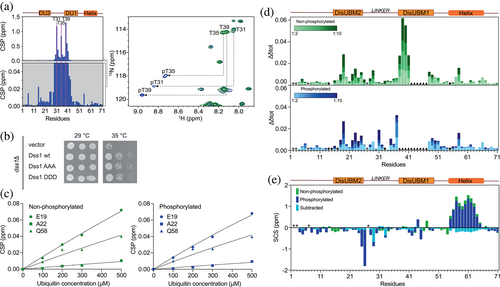
To identify the precise CK2 phosphorylation sites in Dss1, we followed the reaction by nuclear magnetic resonance (NMR) spectroscopy. For this, the resonances of Dss1 were assigned under the used conditions using a set of triple resonance 3D NMR spectra. The addition of CK2 and ATP resulted in marked spectral changes in the 15N-HSQC NMR spectrum. Of particular notice, three distinct peaks appeared in the spectral region where phosphorylated threonines are expected to occur. The resonances of phosphorylated Dss1 were re-assigned after completion of the reaction and this confirmed T31, T35, and T39 as the phosphorylated residues (Figure 3a, right). Mapping of the combined N and HN chemical shift perturbations (CSPs) onto the sequence of Dss1 showed that these concentrated to residues surrounding the three threonines positioned within the non-conserved linker region (Figure 3a, left). Finally, to obtain the phosphorylation kinetics and determine the reaction order of the three threonines, the intensity changes of the three appearing peaks from the phosphorylated states and disappearing peaks from the unphosphorylated states were monitored over time. We note that a set of additional peaks linked to intermediates from neighboring effects were observed. Fitting either one phase or two phase exponentials to the data (Table 1), the latter necessary for modeling the sensitivity to simultaneous nearby phosphorylation events, a distinct sequence of events was deduced. T35 and T39 were phosphorylated simultaneously and at a similar rate, whereas T31 was phosphorylated at a slower rate, with incomplete reaction within the timeframe of the experiment (69 h; Figure S1a). Thus, the linker of Dss1 has three phosphorylation sites targeted by CK2: T35 and T39 are two major sites, while T31 is a minor site. To further analyze the importance of these sites, we tested if overexpression of Dss1 variants could complement the temperature sensitive growth defect of the Dss1 knock-out strain (dss1Δ). Expression of the GFP-tagged Dss1AAA variant and a phosphomimetic variant with the three consensus threonines exchanged with aspartate (Dss1DDD) both complemented the temperature sensitive phenotype similar to WT Dss1 (Figure 3b). Previous studies (Funakoshi et al., 2004; Jossé et al., 2006; Paraskevopoulos et al., 2014; Tomko & Hochstrasser, 2014) have shown that the phenotype of the dss1 knock-out strain is largely attributed to lack of Dss1 incorporation in the 26S proteasome, the growth assay therefore dominantly reports on this function of Dss1. Accordingly, this suggests that the threonine residues have no, or only minor, impact on Dss1 function in the proteasome. However, we note that Dss1 is overexpressed and that using aspartate as a phosphomimetic also may not capture the full effect of phosphorylation, as described previously (Hendus-Altenburger et al., 2017).
| kfast (s−1) | kslow (s−1) | |
|---|---|---|
| Thr31a | ||
| Thr35 | n.a. | |
| Thr39 | n.a. |
- Note: Phosphorylation kinetics of Thr35 and Thr39 were fitted to a one-phase decay model and Thr31 phosphorylation to a two-phase decay model; a fast phase (kfast) sensing the phosphorylation of the Thr35 and Thr39, and a slow phase (kslow), which represents its own phosphorylation.
- a Saturation not achieved.
2.3 Phosphorylation of Dss1 sustains ubiquitin binding but destabilizes its C-terminal helix without unwinding it
Since yeast growth assays suggested intact proteasomal Dss1 function when the three threonines were mutated to alanine, we asked whether triple-phosphorylated Dss1 (3p-Dss1) would bind ubiquitin. Using NMR to follow the binding of mono ubiquitin to 15N-Dss1 and 15N-3pDss1, and mapping of CSPs and peak intensity changes, we observed essentially no differences between nonphosphorylated Dss1 and 3p-Dss1 (Figure 3c,d and Figure S2). Both DisUBMs engaged in binding with ubiquitin and highly similar binding curves were observed for the perturbed residues (Figure 3c), suggesting an affinity in the high micromolar range as shown previously for nonphosphorylated Dss1 (KD = 380 ± 40 μΜ; Dreier et al., 2022). Thus, despite the linker region being phosphorylated, similarly affected residues and binding curves were observed, suggesting that the low affinity for mono-ubiquitin was unchanged by linker modifications (Figure 3c,d and Figure S2).
We next asked if phosphorylation would affect the overall structural properties of Dss1. We therefore assigned Dss1 and 3p-Dss1 in 8 M urea to obtain intrinsic random coil reference values (Modig et al., 2007) to account for the chemical effects of the three consecutive phosphorylations. Using these independently assigned reference spectra, we calculated the secondary chemical shifts (SCSs) of the Cα, Cβ, and C′ nuclei of both Dss1 and 3p-Dss1 and plotted these on a per-residue basis (Figure S1b). The phosphorylations of Dss1 only had a small influence on the secondary structure profiles of 3p-Dss1. However, focusing on the C′, subtraction of the two sets of SCSs recorded under identical conditions revealed that the phosphorylations decreased the population of the C-terminal helix with small additional effects on the population of the extended structure upstream from T31 (Figure 3e). This destabilization was not caused by effect on the helix dipole and we speculate that the reason for this comes from the introduction of several negative charges that will expand the linker region, interfering with the open-close equilibrium between the helix and the linker shown earlier (Schenstrøm et al., 2018). Nonetheless, even minor changes in helical populations in IDPs have been shown to impact normal function (Newcombe et al., 2022) as well as play roles in disease (Chhabra et al., 2018).
2.4 3p-Dss1 binds to the FHA domain of Dma1 using its phosphorylated linker
As the phosphorylations appeared to alter helicity and therefore likely affect the access to the linker region due to the fold-back mechanism of the transient helix, we revisited the reported interaction partners of Dss1 identified by MS (Schenstrøm et al., 2018). In this list, we searched for interaction partners of Dss1 which bind Dss1 independently of the helix. To locate a potential interaction partner for 3p-Dss1, we analyzed the sequence properties of the phosphosites within the linker region of Dss1 and searched the eukaryotic linear motif database (elm.eu.org) for potential motifs. Besides confirming the CK2 phosphorylation motifs, all three threonines of the linker were predicted to reside in motifs interacting with FHA domains, with the consensus sequence pTxx(ED) (Durocher et al., 2000). The FHA domain specifically recognizes phosphothreonine-containing motifs, and not phosphoserines, and is especially prevalent in nuclear proteins that are involved in the cell cycle, DNA repair, and transcriptional regulation (Almawi et al., 2017; Brooks et al., 2008; Durocher et al., 2000); functions also ascribed to the Dss1 portfolio (Kragelund et al., 2016). A subset of FHA interacting ligands have, instead of an acidic n + 3 residue, a large hydrophobic residue, exploiting the consensus sequence of pTxx(IVL) (Durocher et al., 2000). For S. pombe Dss1, all three phosphothreonines confer to the acidic motif.
In the list of previously identified interactors for Dss1 (Schenstrøm et al., 2018), we decomposed the domain composition of proteins for which at least a twofold increase in the amount of pulled-down partner was observed when the helix of Dss1 was removed. Only a single FHA-containing protein was identified. A fourfold increase in the interaction with Dma1 was observed (Schenstrøm et al., 2018), and hence we hypothesized that this was due to an increased access to the linker region and to the three phosphorylated threonines. The FHA domain of Dma1 (residues 25–160) was expressed recombinantly in Escherichia coli and its fold integrity was assessed by far-UV circular dichroism (CD) spectroscopy (Figure 4a). As expected from the structure of FHA domains (Almawi et al., 2017; Reinhardt & Yaffe, 2013), the far-UV CD spectrum confirmed a folded β-sheet structure with an ellipticity minimum at 216 nm. However, the CD spectrum also deviated from a canonical profile expected for a β-sheet fold. As for many other β-sandwich structures, for example, fibronectin type III domains (Dagil et al., 2012), the CD spectrum of the Dma1-FHA domain was imprinted by aromatic exciton couplings, as shown by the distinct lineshapes of the CD spectrum. We then prepared 15N-labeled 3p-Dss1 and added Dma1-FHA to the sample to observe if an interaction occurred. Since the conditions that were needed to stabilize the Dma1-FHA domain differed from those used for the assignments of 3p-Dss1, the NMR spectra of 3p-Dss1 were reassigned to allow for interaction site mapping, and hence the spectrum is slightly different from the one in Figure 2d. Once added to 15N-3p-Dss1, the Dma1-FHA domain induced minor shift changes in the HSQC spectrum, but several resonances completely disappeared, thereby confirming a relevant interaction between 3p-Dss1 and Dma1-FHA with an affinity in the sub-micromolar range (Figure 4b). Mapping of the spectral changes to the sequence of 3p-Dss1 showed clearly that the three phosphorylated threonines all engaged in binding, but also that the affected region stretched out to include mainly DisUBM1 and to some extent DisUBM2, thereby masking these binding sites (Figure 4c). This was further supported by the addition of ubiquitin to the 3p-Dss1-FHA sample, which had no effect of the peak position or intensities (Figure S3b). Interestingly, residues at the C-terminal, including those constituting the α-helical region of 3p-Dss1, were left unperturbed and available for binding (Figure 4b, marked residues). Importantly, the addition of Dma1-FHA to nonphosphorylated Dss1 had only minor effects on the NMR spectrum, suggesting a large decrease in affinity for Dma1-FHA (Figure S3). To support the complex formation shown by NMR, we analyzed 3p-Dss1 and the 3p-Dss1:Dma1-FHA complex using analytical size exclusion chromatography (SEC) and saw a shift in elution volume of 3p-Dss1 eluting alone at 9.85 mL and the complex at 9.36 mL (Figure S4a). Alone, Dma1-FHA had affinity for the column material and eluted after the salt. Finally, we quantified the interactions between 3p-Dss1 and Dma1-FHA using micro-scale thermophoresis, labeling 3p-Dss1 with FITC in the C-terminal cysteine. Although the solubility of the Dma1-FHA domain prohibited measurements at the highest concentrations affecting the precision, we obtained a KD of 1.3 ± 1.1 μM (Figure S4b), again supporting a low-micromolar affinity indicated from the NMR titrations and co-elution on SEC.
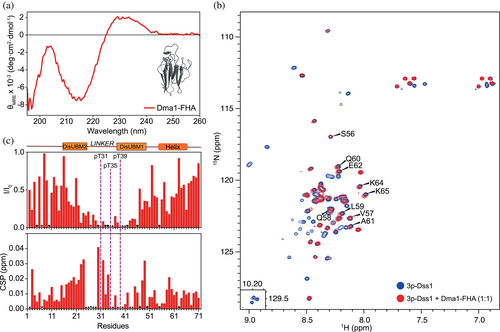
In conclusion, triple phosphorylated Dss1 binds to the FHA domain of Dma1 in a way that masks the ubiquitin-binding sites, primarily DisUBM1, and which leaves the conserved α-helix available for binding to other targets.
2.5 Phosphorylation of Dss1 is evolutionarily restricted
The Dma1 E3-ligase is not conserved in evolution and different species contain different variants (Chahwan et al., 2013). Phylogenetic analyses of FHA-RING ligases have shown that the human E3 ligases checkpoint with forkhead and RING finger domains protein (CHFR) and RING finger protein 8 (RNF8) are more similar to each other than to any of the fungal FHA-RING ligases (Brooks et al., 2008). Furthermore, it has been shown that evolutionarily, the human RNF8 and not CHFR is the likely homolog of S. pombe Dma1 (Chahwan et al., 2013). We therefore reason that the phosphorylation of Dss1 is unlikely to be a conserved feature.
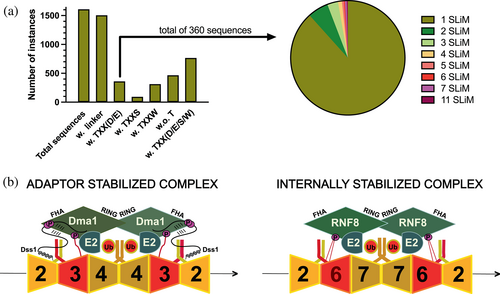
To address this, we first investigated whether human Dss1 would be a substrate of CK2. Despite extensive incubation of human Dss1 with CK2 performed in parallel with S. pombe Dss1, which was readily phosphorylated, we observed no phosphorylation events (Figure S5). This was also expected as human Dss1 does not have any threonines in its linker region, which is shorter than many of the fungal Dss1 proteins. Next, we analyzed the linker region properties in a large set of Dss1 sequences, focusing on the presence or absence of threonines and whether or not any threonine would be in a sequence context that would allow for (i) CK2 phosphorylation and (ii) subsequent FHA interactions. We collected all full-length Dss1 sequences from PFAM/InterPro – entry PF05160 (Figure 5a). Of the 1612 total sequences, we first identified those with a linker region between the two DisUBMs capped by the FxxF on the N-terminal side and by the WxxxW on the C-terminal side. Most (96%; 1506) sequences contained a linker region. Of the 1506 sequences, 360 sequences contained the Txx(D/E) motif, with the majority carrying just one instance of it (318 sequences). However, several sequences had, similar to Dss1, multiple motifs, and one sequence had 11 of these motifs in a very long linker (Figure 5a). We also noticed that other threonine-based motifs were frequent in the linker regions, especially TxxS (94 sequences; e.g., Ustilago maydis) and TxxW (312 sequences; e.g., Amphimedon queenslandica). None of these are known short linear motifs (SLiMs). A total of 466 sequences did not contain any threonines. Finally, we built an evolutionary tree of the seed sequences of PF05160 (Figure S6). From this, it appeared that the presence of the Txx(D/E) motif was not systematic but appeared sporadically. However, once the motif appeared, it would remain in that branch of the tree, and also disappear again, supporting the idea of ex nihilo acquisition of function in IDPs (Davey et al., 2015). This mechanism of ex nihilo SLiM evolution suggests that SLiMs can be acquired by mutations, insertions, and deletions in disordered regions, whereby the evolution of a novel SLiM from “nothing” can occur from the appearance of a functional module to a previously nonfunctional region of a protein sequence. For the phosphorylation and corresponding FHA interaction motif in S. pombe Dss1, this appears to be the most likely case, and may also underlie the different lengths of the linker.
3 DISCUSSION
As a multifunctional IDP, Dss1 already has a long list of interaction partners. Through these, it serves various roles as an assembly factor, a scaffold, and a stabilizer of complexes. All these roles have been assigned to a non-posttranslationally modified Dss1, although it is possible that some of these roles are linked to different posttranslational states. Here, we add to the complexity of the role of Dss1, identifying it as a target of CK2 and as a phosphorylation-dependent interaction partner for the E3-ligase Dma1 in vitro. To our knowledge, none of the identified sites have been observed by phosphoproteomics. Since Dss1 is small and only contains a few arginine and lysine residues, it is possible that phosphorylation is not detected by bottom-up proteomics, as this technique normally relies on trypsin for digestion. However, we also note that since we do not have antibodies recognizing endogenous Dss1, we rely on overexpressed GFP-tagged Dss1 for the yeast studies, and as such, we can currently only speculate on the potential cellular role of the Dss1 phosphorylation and interaction with Dma1.
Fission yeast Dma1, budding yeast Dma1 and Dma2, as well as mammalian RNF8 are members of the unique FHA-RING domain protein family of E3 ligases and are all linked to mitotic regulation and septin organization (Chahwan et al., 2013; Chen et al., 2021; Cullati & Gould, 2019; Fraschini et al., 2004). In S. pombe, Dma1 functions to prevent mitotic exit and cytokinesis during spindle checkpoint arrest by inhibiting septation initiation network signaling (Krapp & Simanis, 2014). Recent MS and pull-down studies have shown that S. pombe Dss1 interacts with all four mitotic septins (Schenstrøm et al., 2018). This interaction, however, was distinctly dependent on the C-terminal helix of Dss1, as its removal abolished binding to all four septins (Schenstrøm et al., 2018). Our NMR studies showed that this helix, albeit slightly destabilized, but not unwound, is fully accessible in complex with the FHA domain of Dma1, suggesting a role of S. pombe Dss1 as an adaptor protein, linking Dma1 to the septins. However, importantly, Dma1-septin interactions are likely to occur also independently of Dss1. Hence, similar to the role of Dss1 in other protein complexes, including the 26S proteasome, the possible function of Dss1 may be to fine-tune assembly and/or stabilize the complex. Perhaps for this reason, the propensity of phosphorylation in the linker regions is not conserved, and human Dss1 does not carry any threonines in its linker, thus not qualifying as a substrate for CK2. One explanation may lie in a similar sequence divergence of the septins as well as the Dma1 orthologs. Although septins have a conserved domain structure with a small GTP-binding (G-) domain, an N-terminal domain, and a C-terminal coiled-coil domain (Cavini et al., 2021), they also contain disordered regions, whose length varies and increases from Spn4 in S. pombe to SEPT9 in humans.
To understand the potential origin of these evolutionary differences, we focused on the role of Dma1. As an E3-ligase, Dma1 controls septin dynamics (Chahwan et al., 2013), recruiting E2s to the septin filament to promote ubiquitylation of septins, shown recently for SEPT7 in humans (Chahwan et al., 2013). In S. pombe, Dss1 has been shown to bind to all mitotic septins (Spn1–4) and do so in a helix-dependent manner (Schenstrøm et al., 2018). Since this interaction may not be direct, this suggests that septins may constitute the link between Dss1 and Dma1. Sequence analysis of septins from humans and S. pombe reveals that the disordered regions of many septins also carry the Txx(D/E) motif (Figure S7). They form heterooligomeric complexes, assembling into filaments that can be organized into higher-order structures. In the mitotic complexes, Spn1, Spn2, Spn3, and Spn4 form an oligomer in S. pombe, whereas Sept6/11, SEPT7, SEPT2, and SEPT9 form an oligomer in humans (Versele & Thorner, 2005). Remarkably, Spn1 and Sept6 (and Sept11), similarly positioned in the oligomers, both carry the Txx(E/D) motif, while in S. cerevisiae, Cdc3 takes on this position and also carries the motif. We speculate that these motifs can be phosphorylated and that depending on the stability of the complex and the Dma1 of the particular species, Dss1 may have evolved to assist in the formation or prevention of this complex. As illustrated in Figure 5b, there can be different ways for Dma1/RNF8 to be linked to the septin oligomers. For instance, accessory proteins such as Dss1 could stabilize the complex (adaptor-stabilized), and/or the complex could be internally stabilized through the disordered regions of the septins (internally stabilized), or both.
FHA-containing proteins have strict specificity for phosphothreonine and FHA domains are relatively rare, and are, according to InterPro found only in 8 and 109 proteins in S. pombe and human proteomes, respectively, playing roles in signal transduction, cell cycle regulation, DNA damage control, and cell growth (Almawi et al., 2017; Brooks et al., 2008; Durocher et al., 2000; Reinhardt & Yaffe, 2013). In FHA domains, only five conserved residues define two binding pockets, one for the phosphorylated threonine (n) and one for the n + 3 residue, which is either a negatively charged D/E or a large hydrophobic residue (Durocher et al., 2000). Recent studies have highlighted that the context surrounding short linear motifs is important for affinity and selectivity (Bugge et al., 2020; Palopoli et al., 2018; Prestel et al., 2019). Similar observations have been made for FHA binding, where ligands can be discriminated through additional interaction surfaces distant to the pockets (Pennell et al., 2010). FHA domains also bind multiphosphorylated sites, one example being Rad53 of budding yeast, which has two threonine phosphorylation sites separated by a single residue, but lacking the conserved acidic n + 3 residue. In the complex with Dun1, the second phosphorylated threonine occupies the canonical site, whereas the first phosphorylation site interacts with the arginine that normally stabilizes the interaction through ionic interactions with the negative charge in the n + 3 position. Indeed, two phosphorylated sites increase the affinity for Dun1 100-fold compared to just one site (Lee et al., 2008). Dss1, but also several septins (e.g., SEPT8, SEPT11, SEPT14, and Spn1) have more than one threonine phosphorylation site located in disordered regions and hence more than one potential interaction site for Dma1. There can be several reasons for this. First, as for Rad53 binding to Dun1, it is possible that an increased affinity may be needed between Dss1 and Dma1, and hence more phosphorylation sites are relevant. As for Rad53, the two main sites in Dss1 (T35 and T39) are also just separated by a single residue. For the septins, the distance can be much larger, which suggests a different mechanism, just as the third phosphorylation site in Dss1 may play a different functional role. It has been shown that increasing the number of phosphorylation sites targeting the same binding site increases partner affinity through allovalent effects (Erlendsson & Teilum, 2021; Mittag et al., 2008; Olsen et al., 2017). The three sites in Dss1, which are located between the two ubiquitin-binding sites, may be necessary to allow dynamic binding and enable FHA to occlude both DisUBMs in Dss1. In this way, Dss1 may not interfere with the ubiquitylation process of the E2 and E3 enzymes. A similar effect may be relevant in septins.
In the original work on the interactions of human Dss1 with RPA, residues in the linker region were key to the interaction. Human Dss1 is not a target for CK2 phosphorylation and lacks threonines in the linker. Instead, the threonines are substituted for either negatively charged residues (spT31(hE32); spT35(hD33)) or a histidine (spT39(hH38)). Substituting eight residues in the linker of human Dss1 (seven negatively charged residues and one tryptophan) to alanine resulted in a strongly impaired interaction with RPA (Zhao et al., 2015). In S. pombe, Dss1 interacts with RPA (Ssb1, Ssb2, or Ssb3), but its linker is already negatively charged (net charge −5; Schenstrøm et al., 2018). However, it is possible that phosphorylation of S. pombe Dss1 affects RPA interactions by increasing the lifetime of the interaction or decreasing the affinity for other competing partners. This remains to be addressed.
In conclusion, we have identified three nonconserved phosphorylation sites in S. pombe Dss1 that are necessary for its interaction with the FHA domain of the E3-ligase Dma1 in vitro. Although we cannot exclude the specificity effect on binding ubiquitin chains, phosphorylation of Dss1 had no influence on mono-ubiquitin binding. Instead, binding of the phosphorylated Dss1 to the FHA domain of Dma1 covered the disordered ubiquitin-binding motifs, but left the C-terminal helix accessible for binding. We speculate that phosphorylated Dss1 could act as an adaptor protein stabilizing the complex between Dma1 and septins, where the linker tethers to the FHA-domain of Dma1 and the helix to the septins. Remarkably, the Txx(D/E) motif is also present in the disordered regions of a subset of septins, suggesting other ways to stabilize the complex, which may depend on the species and the exact septin oligomer organization. We found that the Txx(D/E) motif was nonconserved through evolution but appeared sporadically in 25% of the sequences supporting an ex nihilo appearance of the trait. We additionally discovered two other threonine-based motifs prevalent in many Dss1 linkers with a so far unknown function. Our work expands the interaction portfolio of Dss1 and supports the ex nihilo acquitions of function in IDPs.
4 MATERIALS AND METHODS
4.1 Protein purification
S. pombe Dss1 was expressed and purified as described, either as a GST-tagged version (Schenstrøm et al., 2018) or with a hexa-histinde tag and a SUMO tag in its N-terminal (Newcombe et al., 2021) and human Dss1 as in Ruidiaz et al. (2021). The GST-tagged S. pombe Dss1 variant carried five N-terminal amino acids, being remnant cleavage site residues (GPLGS), and the hexa-histidine-SUMO-tagged variant carried an Asn to Cys substitution in the C-terminal. These two proteins are referred to in the paper as Dss1.
For the FHA domain of S. pombe Dma1, the DNA sequence corresponding to the region 25–160 was inserted into a modified pET24a vector (Twist), adding a hexa-histidine tag and a SUMO tag to its N-terminal, both of which can be cleaved off with ubiquitin-like protein protease 1 (ULP1). The protein was expressed in E. coli NiCo21(DE3) cells using Luria-Bertani media with 50 μg/mL kanamycin. The cells were grown at 37°C while shaking (180 rpm) to an OD600 of ~0.8 at which protein expression was induced with 0.1 mM isopropylthiogalactopyranoside for ~20 h at 18°C. Cells were collected by centrifugation at 5000g, 20 min, 4°C, and the pellet was resuspended in lysis buffer (20 mM HEPES [pH 8], 200 mM NaCl, 20 mM Imidazole, and 5 mM β-mercaptoethanol) supplemented with 1 cOmplete™ EDTA-free Protease Inhibitor Cocktail tablet (Roche) and lysed using a French pressure cell disrupter (Constant Systems) at 20 kpsi. The lysate was supplemented with 1 mM MgCl2 and 125 U Benzonase® (MERCK) and incubated on ice for 30 min to remove DNA before the lysate was cleared by centrifugation at 20,000g for 45 min. The soluble fraction was applied to Nickel Sepharose Fast Flow resin (5 mL; GE Healthcare) which was equilibrated with 10 column volumes (CVs) of lysis buffer. The resin was washed with 10 CV high-salt washing buffer (20 mM HEPES [pH 8], 1 M NaCl, 20 mM Imidazole, and 5 mM β-mercaptoethanol), followed by washing with 10 CV lysis buffer, and the protein was eluted with 3 CV of elution buffer (20 mM HEPES [pH 8], 200 mM NaCl, 250 mM Imidazole, and 5 mM β-mercaptoethanol). The eluate was dialyzed against 2 L of 20 mM HEPES (pH 8), 100 mM NaCl, and 5 mM β-mercaptoethanol at room temperature while cleaving the fusion protein using 25 μg of ULP1. Subsequently, the sample was applied to a HiTrap SP FF column (5 mL; GE Healthcare) and eluted using a linear gradient of 100 mM to 1 M NaCl over 20 CV. The fractions containing Dma1-FHA were pooled and concentrated using a 3-kDa cutoff Amicon® Ultra-15 Centrifugal Filter (Millipore) and applied to a HiLoad 16/600 Superdex 75 pg column (GE Healthcare) equilibrated with 25 mM Tris–HCl (pH 7.5 at 25°C), 150 mM NaCl, and 5 mM β-mercaptoethanol for final purification and buffer change. Analytical SEC was run on a Superdex 75 GL 10/300 column (GE Healthcare) in 25 mM Tris (pH 7.5), 150 mM NaCl with a flow rate of 0.8 mL/min.
4.2 Bioinformatics
Prediction of phosphorylation sites and associated kinases were performed using NetPhos (Blom et al., 1999), NetPhosK (Blom et al., 2004), NetPhosYeast (Ingrell et al., 2007), GPS3.0 (Wang et al., 2020), Scansite 3 (Obenauer et al., 2003), and Phospho.ELM 6.0 database using standard setting. Mammalian sites specific for five different kinases (PKA, PKC, PKG, CK2, and CaM-II) were included in the training set. Full-length sequences were extracted from PFAM/Interpro (El-Gebali et al., 2019; Mitchell et al., 2019; Punta et al., 2012) entry PF05160 and analyzed for the presence of threonine motifs. A phylogenetic tree associated with the PFAM seed alignment was extracted from the same source. The tree was annotated with threonine matches and species information from UniProt (UniProt Consortium, 2023) and visualized using the ete3 python library (Huerta-Cepas et al., 2016).
4.3 Phosphorylation assays
CK2 from proteinkinase.de (lot 080708) was used. Initial in vitro phosphorylation experiment conditions were optimized as described (Liokatis et al., 2012; Selenko et al., 2008), including varying temperature, substrate concentration, and amount of kinase. The initial experiments were performed in 25 mM Tris–HCl pH 7.5, 50 mM NaCl, 20 mM MgCl2, 5 mM ATP, 5 mM DTT, and varying amounts of active CK2 holoenzyme.
15N-Dss1 used for FHA interaction analyses was phosphorylated using 1.5 μg active CK2 kinase pr. mg protein (lot 656154/08). The phosphorylation experiment was performed in 50 mM Tris–HCl (pH 7.5 at 5°C), 50 mM NaCl, 20 mM MgCl2, 10 mM ATP, and 5 mM DTT. The reaction was incubated at 25°C for 48 h. A control sample was treated identically, however absent of CK2 kinase. Phosphorylation was followed by SDS-PAGE, MALDI-TOF MS, and NMR spectroscopy. The final conditions for phosphorylation followed by NMR are indicated below.
4.4 CD spectropolarimetry
4.5 Microscale thermophoresis
3p-Dss1 was labeled with 5-Iodoacetamidofluorescein (5-IAF; Thermo Fisher Scientific) in phosphate-buffered saline (PBS) buffer pH 7.5 with 6 M GuHCl and the reaction run for 4 h in the dark at room temperature. Excess 5-IAF was removed by dialysis toward 50 mM NH4HCO3 overnight, followed by RP-HPCL using a SOURCE 15RPC ST 4.6/100 column and 50 mM NH4HCO3 with a gradient of 0%–70% acetonitrile. The protein concentration was determined from the absorbance at 492 nm using ε = 82,000 M−1 cm−1. Thermophoresis was measured with a 3p-Dss1 concentration of 40 nM and increasing concentrations of added Dma1-FHA following the protocol from Chhabra et al. (2023).
4.6 MALDI-TOF mass spectrometry
MALDI-TOF MS matrix and samples were prepared on target via dried droplet method by embedding 1 pmol of Dss1 in a saturated solution of 4-hydroxy-α-cyanocinnamicacid (HCCA) matrix. Measurements were performed using a Bruker Autoflex. External calibration was performed using the (M + H) + ions of a PS1 peptide mixture from GE Health Care Life Sciences.
4.7 NMR experiments
S. pombe Dss1 was assigned in nonphosphorylated and triple-phosphorylated states under native conditions and in 8 M urea for internal referencing. Spectra of all nonurea samples were recorded on a 600 MHz Bruker spectrometer, while spectra of urea samples were recorded on a 750 MHz Varian spectrometer. All spectra used for assignment were recorded at 5°C and all 3D spectra (Bruker Biopack) were recorded using nonlinear sampling with a 25% data reduction.
4.7.1 Phosphorylation kinetics
A sample of 250 μM 15N labeled Dss1 in identical conditions as used for backbone resonance assignment was prepared. The experiment was performed at 25°C and a series of 1H,15N HSQC spectra were recorded at temperatures of 5°C, 10°C, 15°C, 20°C, and 25°C to allow for reliable and unambiguous resonance assignment. A similar temperature series was recorded for phosphorylated Dss1. An aliquot of 5 μL 0.3 μg/μL active CK2 was added to the sample, which was mixed carefully and immediately inserted into the spectrometer. A series of 51 successive HSQC spectra were recorded over a time period of approximately 42 h with two additional HSQC spectra recorded at approximately 53 and 68 h after kinase addition. Phosphorylation kinetics was extracted by fitting to either single or double exponential decay.
4.7.2 Ubiquitin binding
Four stock samples were prepared from two stock samples of 250 μM unphosphorylated and phosphorylated 15N labeled Dss1, respectively, with identical buffer composition of 25 mM Tris–HCl (pH 7.5), 50 mM NaCl, 5 mM DTT, 20 mM MgCl2, and 10 mM ATP. Ubiquitin-binding stock samples A and B contained 50 μM phosphorylated Dss1 and either no or 500 μM mono ubiquitin (H. sapiens) purchased from Sigma-Aldrich. Ubiquitin binding stock samples C and D contained 50 μM nonphosphorylated Dss1 and either no or 500 μM mono ubiquitin. A series of five HSQC spectra were recorded in titration series of phosphorylated and unphosphorylated Dss1 with mono ubiquitin at constant Dss1 concentration and ubiquitin concentrations of 0, 100, 200, 300, and 500 μM by mixing the stock samples A and B, and C and D, appropriately. Changes in chemical shift as a function of ubiquitin concentration were fitted to a one-site binding site to extract an apparent KD, as described (Teilum et al., 2017).
4.7.3 FHA interaction analyses
Samples of 50 μM of both 15N-Dss1 and 15N-3p-Dss1 with and without the addition of Dma1-FHA (1:1) were prepared from the phosphorylation assays described above. Final experimental conditions were 25 mM Tris–HCl (pH 7.5 at 5°C), 50 mM NaCl, 5 mM DTT, 125 μM 4,4-dimethyl-4-silapentane-1-sulfonic acid, and 5% D2O. Additionally, approximately 6.5 mM MgCl2 and 1.5 mM ATP remained in the samples from the phosphorylation assays. 15N-best-HSQC spectra of 15N-Dss1 and 15N-3p-Dss1 in the absence and presence of Dma1-FHA were recorded at 5°C to monitor interactions. All spectra were recorded on Bruker Avance III HD 750 MHz spectrometer equipped with a cryogenic probe using Bruker Topspin (v.3.6.0). Binding of ubiquitin to a preformed 1:1 complex of 15N-3p-Dss1 was measured in a 15N-HSQC spectrum following the addition of ubiquitin in a 1:1:1 ratio recorded on a 600 MHz Bruker Avance III HD spectrometer.
4.7.4 Data processing
4.8 Yeast strains and methods
The dss1Δ (dss1::G418) strain was kindly provided by Dr Colin Gordon. Standard genetic methods and media were used (Moreno et al., 1991). The plasmids used for overexpression of dss1 were in the pDUAL vector carrying ura4+ for selection and the nmt1 promoter, as described before (Paraskevopoulos et al., 2014). The Dss1AAA (T31AT35AT39A) and Dss1DDD (T31DT35DT39D) expression constructs were purchased from Genscript. Phospho-labeling and purification of GFP-tagged Dss1 were performed largely as described previously (Benito et al., 1998; Schenstrøm et al., 2018). Briefly, the cells expressing GFP or GFP-tagged Dss1 were grown at 29°C for 12 h in phosphate-free minimal media (EMM) containing 50 μM NaH2PO4 and supplements. At a cell density of 5 × 106 cells/mL, 10 mL culture was harvested, washed in water by centrifugation, and resuspended in 10 mL fresh media containing 5 mCi P-32 orthophosphate. The cells were then incubated for 6 h at 29°C. Lysates were prepared using glass beads in 50 mM Tris–HCl (pH 7.5), 100 mM NaCl, 10 mM EDTA, Complete protease inhibitors (Roche), and phos-stop (Roche). SDS was added to 1% and the extracts were diluted 10-fold in RIPA buffer (10 mM NaH2PO4 [pH 7.0], 150 mM NaCl, 10 mM EDTA, 1% Triton X-100, and phos-stop [Roche]). The lysates were cleared by centrifugation (13,000g, 30 min.), and the supernatants were used for immunoprecipitation using GFP-trap (Chromotek) beads. After 4 h end-over-end tumbling at 4°C, the beads were extensively washed in RIPA buffer and protein eluted with SDS sample buffer (63 mM Tris–HCl [pH 6.8], 2% SDS, 25% glycerol, 0.01% bromphenol blue, and 5% β-mercaptoethanol). The samples were resolved on 12.5% SDS-PAGE gels, fixed in destain (50% ethanol and 10% acetic acid), dried, and exposed at −80°C to x-ray film (Agfa) with intensifying screens. As a reference, GFP and the GFP-tagged Dss1 variants were purified from unlabeled 1 L cultures as above. The proteins were separated on 12.5% SDS-PAGE gels and stained with Coomassie Brilliant Blue. Growth assays were performed by spotting serial dilutions of exponential phase cultures on solid media, as described previously (Schenstrøm et al., 2018).
AUTHOR CONTRIBUTIONS
Nina L. Jacobsen: Formal analysis; investigation; visualization; writing—original draft; writing—review and editing. Magnus Bloch: Formal analysis; investigation; visualization; writing—review and editing. Sarah F. Ruidiaz: Formal analysis; investigation. Peter Millard: Formal analysis; investigation. Jonas D. Elsborg: Formal analysis; investigation. Wouter Boomsma: Formal analysis; investigation. Ruth Hendus-Altenburger: Formal analysis; investigation, supervision. Rasmus Hartmann-Petersen: Conceptualization; funding acquisition; resources; supervision; writing—review and editing. Birthe B. Kragelund: Conceptualization; funding acquisition; investigation; project administration; resources; supervision; validation; writing—original draft; writing—review and editing.
ACKNOWLEDGMENTS
The authors thank Signe A. Sjørup and Anne-Marie Lauridsen for expert technical assistance and Dr Andreas Prestel for NMR support. This work was made possible by the Novo Nordisk Foundation Challenge grant REPIN—rethinking protein interactions (#NNF18OC0033926 to BBK and RHP). The authors thank Villum Fonden and the Novo Nordisk Foundation for support for NMR infrastructure. NMR data were in part recorded at cOpenNMR—an infrastructure facility funded by the Novo Nordisk Foundation (#NNF18OC0032996). The work was also supported by the Novo Nordisk Foundation through the MLLS Center for Basic Machine Learning Research in Life Science (#NNF20OC0062606).