Conformational landscape and pathway of disulfide bond reduction of human alpha defensin
Abstract
Human alpha defensins are a class of antimicrobial peptides with additional antiviral activity. Such antimicrobial peptides constitute a major part of mammalian innate immunity. Alpha defensins contain six cysteines, which form three well defined disulfide bridges under oxidizing conditions. Residues C3-C31, C5-C20, and C10-C30 form disulfide pairs in the native structure of the peptide. The major tissue in which HD5 is expressed is the crypt of the small intestine, an anaerobic niche that should allow for substantial pools of both oxidized and (partly) reduced HD5. We used ion mobility coupled to mass spectrometry to track the structural changes in HD5 upon disulfide bond reduction. We found evidence of stepwise unfolding of HD5 with sequential reduction of the three disulfide bonds. Alkylation of free cysteines followed by tandem mass spectrometry of the corresponding partially reduced states revealed a dominant pathway of reductive unfolding. The majority of HD5 unfolds by initial reduction of C5-C20, followed by C10-C30 and C3-C31. We find additional evidence for a minor pathway that starts with reduction of C3-C31, followed by C5-C20 and C10-C30. Our results provide insight into the pathway and conformational landscape of disulfide bond reduction in HD5.
Introduction
Animals and plants produce a variety of antimicrobial peptides.1 These antimicrobial peptides are produced in epithelial tissues, thus contributing to the barrier that it forms against invading microbes and viruses.2, 3 In addition to their antibiotic activity, antimicrobial peptides often also modulate immune activity.4 They are an integral part of mammalian innate immunity and contribute to the first line of defense against infectious disease. By virtue of both their antimicrobial and immune modulating activities, various classes of antimicrobial peptides are being developed for use as peptide therapeutics in the hope to meet the ever growing requirement for new antibiotics.5 Peptide therapeutics may often carry cysteine residues that are paired in disulfide bonds, which may play a key role in oxidative folding of the peptide, stabilization of the peptide fold, provide potential sites for functionalization, and confer enhanced stability against proteases.6
Alpha defensins are a class of antimicrobial peptides that are rich in arginines and hydrophobic residues and additionally carry six cysteine residues that form the characteristic disulfide bridge structure of the peptide.2 In addition to killing both Gram-positive and Gram-negative bacteria, as well as fungi and protozoa, human alpha defensins also have broad antiviral activity.7 Human alpha defensin 5 (HD5) in particular has known antiviral activity against adenovirus, human papilloma virus, herpes simplex virus, and polyomavirus. HD5 is mainly produced by Paneth cells in the crypts of the small intestine.2 It is translated as a 94 amino acid long precursor that is processed by trypsin into the 32 amino acid long final product.8 The peptide has six cysteine residues that form three disulfide bridges: C3-C31, C5–20, and C10-C30 (see Fig. 1).9, 10 These disulfide bridges are crucial to form the characteristic fold of alpha defensins, which consist of a three-stranded anti-parallel beta-sheet that is connected via a coil between strand 1 and 3, and a loop between strand 3 and 2. The disulfide bridge C3-C31 connects the peptide near the two termini on strand 1 and 2, C5-C20 connects strands 1 and 3, while C10-C30 connects strand 2 to the coil between stands 1 and 3.
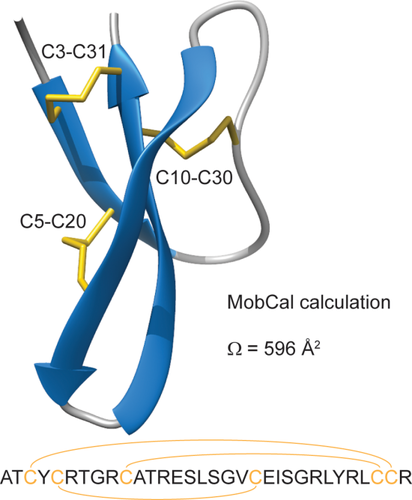
Sequence and crystal structure of human alpha defensin 5, highlighting the disulfide bridges (PDB code: 1zmp). The calculated collisional cross section (Ω) of HD5 according to the MobCal Trajectory Method is listed.
Intact disulfide bridges are crucial for some, but not all of HD5's antimicrobial or antiviral activity.11-13 Interestingly, human beta defensins—which have a similar characteristic disulfide bridge structure—require disulfide bond reduction for efficient antimicrobial activity.14 Reduction of all three disulfide bridges of HD5 actually results in a strong zinc ion chelating peptide.15 On one hand, zinc ion chelation is thought to be the mechanism of action of some antimicrobial peptides like calprotectin.16 On the other hand, zinc-binding also confers protease resistance to reduced HD5.15 These reports all point toward a possible physiological role of (partly) reduced HD5. Though speculative, the structural plasticity associated with disulfide bond reduction may contribute to the fact that such a simple 32 amino acid long peptide can display such a wide variety of antimicrobial, antiviral and immune modulating activities.
We investigated the pathway of disulfide bond reduction of HD5 using tandem mass spectrometry and ion mobility spectrometry (IMS) coupled to mass spectrometry (MS; the couple of techniques is referred to as IMMS).17, 18 In ion mobility spectrometry, ions are separated based on differences in shape and charge. Several previous studies suggest that when combined with ESI and MS, ion mobility provides information about the populations of conformations that biomolecular ions adopt coming from solution.19, 20 We found evidence of stepwise unfolding of HD5 as the three disulfide bridges sequentially break. By performing experiments on partially reduced states of HD5, which were alkylated with iodoacetamide and subsequently analyzed by electron transfer dissociation with collisional activation (ETcaD) tandem MS, we were able to demonstrate that reductive unfolding of HD5 follows a defined pathway. Our results shed light on the transition between oxidized and reduced HD5 in terms of the order of disulfide bond reduction and the associated conformational landscape.
Results
We set up a time course of HD5 reduction in 2-mercaptoethanol in the hope to catch partially reduced states of HD5 by IMMS from which we could infer the pathway of disulfide bond reduction. Without addition of reducing agent, fully oxidized human alpha defensin 5 (HD5ox) appeared as a single species with a mass of 3581 ± 1 Da (theoretical mass 3582 Da) and a main charge state of 4+ (see Fig. 2). Note that the peptide was analyzed at a concentration of 5 µM under non-denaturing conditions (5 mM ammonium acetate, pH 5), yet no higher order oligomers were detected. After incubation for >10 min in 10 mM of 2-mercaptoethanol, we observed a mass shift to 3588 ± 1 Da, consistent with full reduction of HD5ox to HD5red (theoretical mass 3588 Da). HD5red also displayed a shift in charge state to 5+, which indicates that the peptide has a larger chargeable surface area compared to HD5ox, possibly as a result of unfolding.21 We incubated stock of HD5ox in 2 mM 2-mercaptoethanol for 10, 20, and 40 min to observe the intermediate steps of this reductive unfolding. Whereas the 5+ charge state displayed a bimodal mass distribution, thereby indicating a fast complete transition from HD5ox to HD5red, the peak shapes of both the 4+ and 5+ charge state in all three time points also deviate from those observed in spectra of HD5ox/HDred, thereby indeed indicating the presence of partially reduced states of HD5.
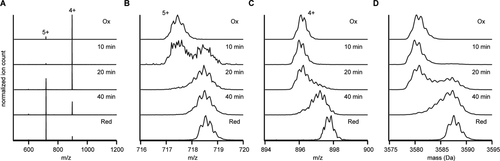
Reduction of HD5 monitored by mass spectrometry. (A) Full range ESI MS spectra of indicated samples, HD5 Ox, Red, and HD5 incubated in 2 mM 2-mercaptoethanol for the indicated times. Charge states are annotated on the spectra. (B) Zoom-in of the 5+ charge state. (C) Zoom-in of the 4+ charge state. (D) Deconvoluted zero charge spectra.
The drift time distribution of HD5ox indicated the presence of at least six gas-phase conformers for the 4+ and 5+ charge states combined [see Fig. 3(A)]. The observed drift times correspond to collisional cross sections of 557 and 565 Å2 (4+) and 584, 593, 615, and 629 Å2 (5+). The main conformers of 565 Å2 (4+) and 584 Å2 (5+) are slightly smaller than those calculated with MobCal based on the crystal structure of HD5 (596 Å2, see Fig. 1).22 The conformational landscape changes dramatically in HD5red. The 4+ charge state shifts to a distribution containing conformers of 579, 590, 601, 612, and 639 Å2, with 612 Å2 the main conformer. The 5+ charge state shifts toward one major conformation of 733 Å2 with one additional minor conformer at 770 Å2. As indicated by the charge state shift of HD5red relative to HD5ox, the consistent increase in collisional cross section of these conformers also indicated unfolding of the peptide following reduction of its disulfide bridges. This is similar to previous IMS observations of reduced lysozyme and bovine pancreatic trypsin inhibitor.23, 24 Removal of the disulfide bridges by point mutations in beta defensin was also shown to result in larger collisional cross sections.25 In the three samples of partially reduced HD5, the peptide populated intermediate collisional cross sections more heavily. As illustrated in Figure 3(B), there is a trend for an increase in m/z with increasing drift time for both charge states. This indicates the presence of partially reduced states of HD5 that favor conformations with larger collisional cross sections. The ion mobility and mass spectrometry data combined clearly demonstrate that initial reduction of the first disulfide bridge results in partial unfolding of HD5, which leads to rapid further reduction to generate the more extensively unfolded HD5red.
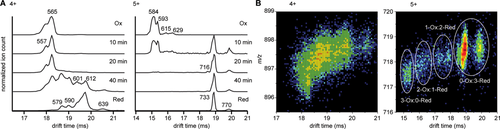
Reduction of HD5 monitored by ion mobility spectrometry. (A) Drift time profile of the 4+ and 5+ charge states, as indicated. Collisional cross sections (Å2) corresponding to the peaks are annotated on the profile. (B) 2D plots of drift time versus m/z.
Considering sequential reduction of the three disulfide bridges of HD5, there is a total of six conceivable pathways between HD5ox and HD5red. We wanted to determine whether there was a defined order in which the disulfide bridges of HD5 are reduced in the presence of 2-mercaptoethanol, or alternatively whether the intermediate states and conformers observed in our IMMS experiments of HD5 reduction represent a mixture of all possible states. Because of the width of the isotope distribution of HD5, the four possible redox states are not sufficiently separated in mass to individually select each state for subsequent tandem MS analysis. Moreover, the mass difference of +1 Da per cysteine residue associated with disulfide bond reduction is so small that any one particular fragmentation pattern might be difficult to assign to any one specific intermediate of HD5 reduction. By alkylating the free cysteines with iodoacetamide, thus adding a carboxyamidomethyl group of +57 Da to each cysteine side chain, we ensure sufficient separation between the four redox states of HD5 as well as introduce an easily distinguishable reporter group to confirm which disulfide bridges are reduced (and alkylated) in the intermediate redox states. Similar to the IMMS experiment, we incubated stock HD5ox in 2 mM 2-mercaptoethanol for 10, 20, 40, and 60 min before quenching the reduction reaction and alkylating free cysteines by the addition of 25 mM iodoacetamide (see Fig. 4). The resulting spectra showed residual HD5ox and three additional species separated a consecutive +114 Da, corresponding to the addition of two, four, and six carboxyamidomethyl groups to HD5. The alkylated species of HD5 are paired with additional species where a 2-mercaptoethanol adduct is present instead of a carboxyamidomethyl group [labeled with an asterisks in Fig. 4(A)]. As 2-mercaptoethanol adducts did not present themselves in our IMMS experiment, we believe they originate from the acid conditions of the desalting procedure and subsequent analysis steps. Presumably, the 2-mercaptoethanol adducts are only transiently formed at higher pH when the reaction rate is high, but locked in place as the reaction rate is lowered under acidic conditions. Regardless, HD5 with two, four, and six carboxyamidomethyl groups represent the main species formed in the reaction, thus allowing us to select the intermediate redox states for tandem MS to determine their disulfide linkage from the +57 Da reporter groups.
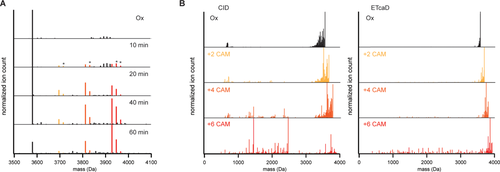
Alkylation of partially reduced states of HD5 and tandem MS analysis. (A) Deconvoluted mass spectra of partially reduced/alkylated HD5. Asterisks indicate species with one or two 2-mercaptoethanol adducts instead of carboxyamidomethyl groups. (B) Deconvoluted tandem MS CID and ETcaD spectra of the indicated precursors. Spectra shown are from 6+ precursor. CAM: carboxyamidomethyl.
HD5ox and HD5 with two, four, and six carboxyamidomethyl groups were analyzed by tandem MS using both collision induced dissociation (CID) and electron transfer dissociation with supplemental collisional activation (ETcaD) as fragmentation techniques. Figure 4(B) shows the deconvoluted tandem MS spectra of all four species. Both fragmentation techniques show an interesting trend in the overall fragmentation efficiency of HD5. Other than a multitude of water and ammonium losses, tandem MS spectra of HD5ox and the two intermediate redox states (plus two or four carboxyamidomethyl groups) show mostly intact precursor and very little backbone fragmentation. However, as all three disulfide bridges are reduced and all six cysteines alkylated, many additional backbone fragments emerge in the tandem MS spectra. What this indicates is that reduction of the first and second disulfide bridge allow for little additional fragmentation compared to HD5ox, suggesting that the final disulfide bridge spans most of the structure of HD5, thus preventing backbone cleavage to yield smaller fragments in the tandem MS spectra. Based on the overall fragmentation efficiency, C3-C31 thus appears to be the last disulfide bridge to reduce in the majority of HD5 in our samples.
Though backbone fragments are low abundant compared to precursor peaks (including water/ammonium losses) in tandem MS spectra of HD5 plus two and four carboxyamidomethyl groups, they are nevertheless detected, especially in the ETcaD spectra. By assigning these fragments to the sequence of HD5, considering variable modification of cysteine with a carboxyamidomethyl group, we were able to distinguish which possible reduction pathways were taken by HD5. As detailed in Figure 5(A), there are three possible forms for both intermediately reduced/alkylated states of HD5. Whereas many fragments will result in identical masses, each scenario does have a number of unique reporter fragments that provide conclusive evidence for the presence of these forms of the peptide [indicated in red in Figure 5(A), matched fragments are listed in Table 1]. Considering all possible scenarios, fragments from ETcaD spectra were assigned to HD5 as shown in Figure 5(B). From tandem MS ETcaD spectra of the first intermediate (with two carboxyamidomethyl groups) we found strongest evidence for initial reduction of C5-C20. We found no evidence for reduction of C10-C30 and minor evidence for initial reduction of C3-C31. In tandem MS ETcaD spectra of the second intermediate (with four carboxyamidomethyl groups) we found evidence of reduction of C5-C20/C10-C30 and of C3-C31/C5-C20. We found no evidence for reduction of C3-C31/C10-C30 in this intermediate.
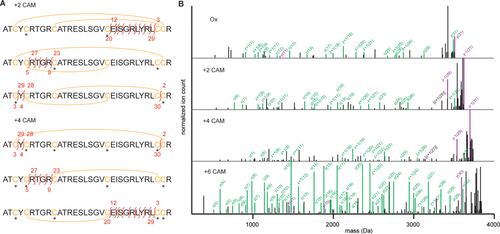
Peak assignments of the ETcaD spectra reveal a dominant pathway of HD5 reduction. (A) Schematic of possible peptide structures corresponding to the selected precursor. Cysteines are highlighted in yellow, disulfide bridges indicated with a yellow arc, carboxyamidomethyl groups on cysteines indicated with an asterisks. The unique backbone fragment ions, or reporter ions, for each possible intermediate are indicated in red. (B) Annotated ETcaD spectra from the 6+ precursors of the indicated states. All peaks bigger than y(31), the largest possible backbone fragment, were removed from the spectra for clarity. Matched peaks are shown for the dominant reduction pathway: in the order C5-C20, C10-C30, C3-C31. Fragments from c/z-related series are indicated in green, b/y-related series in purple. Some notable fragments corresponding to the minor pathway (in the order C3-C31, C5-C20, C10-C30) are indicated in black.
| Intermediate | Identified reporter ions | |
|---|---|---|
| 6+ | ||
| 2_CAM | C5-C20 | z+1(8); z+1(9); z+1(11); z+1(12); c(22); c(23); c(26) |
| C10-C30 | — | |
| C3-C31 | z+1(29); y-1(29) | |
| 4_CAM | C5-C20/C10-C30 | y-1(29) |
| C3-C31/C5-C20 | z+1(27); c(9) | |
| C3-C31/C10-C30 | — | |
| 5+ | ||
| 2_CAM | C5-C20 | z+1(8); z+1(9); z+1(11); z+1(12) |
| C10-C30 | — | |
| C3-C31 | z+1(29) | |
| 4_CAM | C5-C20/C10-C30 | b(30) |
| C3-C31/C5-C20 | c(6); c(8); z+1(24; z+1(25); z+1(27) | |
| C3-C31/C10-C30 | — | |
- CAM: carboxyamidomethyl. The linkages listed under “intermediate” refer to the reduced bonds.
The tandem MS ETcaD spectra of partially reduced/alkylated HD5 thus provide evidence for two pathways of disulfide bridge reduction, based on unique reporter fragments of each possible intermediate state. In the first pathway, the first reduced disulfide bridge is C5-C20, followed by C10-C30 and finally C3-C31. In the other pathway, reduction starts with C3-C31, followed by C5-C20 and C10-C30. In addition to the reporter ions highlighted in Figure 5(A), there are additional fragments that allow us to distinguish between these two established pathways, most notably y/z(29) and b(30) (see Supporting Information Fig. S1). As these could be commonly assigned between the alternative scenarios, we used their relative intensity as a proxy for how much of HD5 reduced via each pathway. From the first intermediate (with two carboxyamidomethyl groups) we estimate that 20 ± 8% (average ± standard deviation from 5+ and 6+ precursor) follows the second pathway (first C3-C31, followed by C5-C20 and C10-C30). Similarly, from the second intermediate we estimate that 19 ± 5% takes the second pathway. Thus, as summarized in Figure 6, our tandem MS ETcaD experiment suggests that the major pathway (∼80%) of disulfide bridge reduction of HD5 start with reduction of C5-C20, followed by C10-C30 and C3-C31.

Major pathway of HD5 reduction. Remaining intact disulfide bridges are listed alongside the schematic structures of the intermediate states. Cysteines are highlighted in yellow, disulfide bridges indicated by yellow arcs, free cysteines labeled with an asterisks.
Discussion
We investigated HD5 by IMMS and tandem MS to reveal the pathway of disulfide bond reduction and explore the conformational landscape associated with the transition from HD5ox to HD5red. We demonstrated that reduction of the initial disulfide bond results in rapid full reduction, as HD5red is the main reduced species throughout the full time course, even at earlier time points. Drift time analysis revealed that HD5red adopts more extended conformations in the gas phase compared to HD5ox, which is likely related to solution-phase unfolding of HD5 upon disulfide bond reduction (as evidenced by the shift to higher charge state). Partially reduced states of HD5 preferentially adopt increased but not fully extended conformations in the gas phase. We find evidence for a major pathway of disulfide bond reduction where first C5-C20, then C10-C30 and finally C3-C31 are reduced. Alternatively, a minor pathway that starts with reduction of C3-C31, followed by C5-C20 and finally C10-C30 is also observed. Hence C5-C20 is most susceptible to reduction with 2-mercaptoethanol, followed by C3-C31, whereas C10-C30 remains relatively unreactive until C5-C20 is reduced.
Pathways of reductive unfolding have previously been reported for Ribonuclease A, bovine pancreatic trypsin inhibitor (BPTI), and alpha lactalbumin.26-28 Similar to our findings for HD5, reductive unfolding of Ribonuclease A was also found to proceed via two alternative pathways. BPTI and alpha lactalbumin, however, only showed evidence for one pathway of reductive unfolding. The latter two cases both presented a single intermediate corresponding to a singly or doubly reduced species for BPTI and alpha lactalbumin, respectively. As pointed out above, reduction of C5-C20 in HD5 appears rate limiting for the further reduction of C10-C30, but not of C3-C31. Based on this limited number of cases it thus seems that no general pathways for reductive unfolding can be formulated at present. The pathway of reductive unfolding seems to be rather protein-specific and might additionally depend on the reducing agent in question.
It was recently reported that HD5 may also exist as a covalently linked dimer that forms spontaneously from HD5ox under aerobic conditions.29 This dimer is in a fully oxidized state, but forms a C5-C20 intermolecular disulfide bridge between the opposing subunits of the dimer, rather than an intramolecular disulfide bridge between stands 1 and 3 within the monomer. The high susceptibility of C5-C20 to reduction provides a rationale for the formation of the covalent dimer. It was additionally reported that mutating out C5-C20 from the sequence of HD5 results in loss of secondary structure of the peptide.11 This might explain the extended conformations of the intermediate reduced species in our IMMS experiment. The integrity of C5-C20 was shown to be crucial for the antimicrobial activity of HD5 against Escherichia coli.11 Residue L29 presents on the same face of the monomer as C5-C20 and was also shown to be crucial for antimicrobial activity of HD5.30 Conversely, residues that are crucial for the antiviral activity of HD5 (R9, L16, V19, L26, and R28), against most notably adenovirus, all present on the opposite face of C5-C20.31, 32
It was shown that a mutant form of HD5 in which all cysteines were replaced by alpha-aminobutyric acids was still active against E. coli but not Staphylococcus aureus (Gram-negative vs. Gram-positive).13 In addition, HD5red has the intriguing property to chelate zinc ions, which follows the antimicrobial mechanism of some other antimicrobial peptides.15, 16 The diverse activity of HD5 against bacteria and viruses seems unusual given the relative simplicity of the peptide. It may be that the transition between HD5ox, HD5red and metal-ion bound states confers a great structural plasticity to the peptide that facilitates the diverse activity of alpha defensins. We have gained new insight into the transition between several key structures of HD5 from our experiments.
Materials and Methods
Sample preparation
Human alpha defensin 5 was purchased from Peptides International (Louisville, KY). The powder was dissolved in MilliQ water to make a 100 µM stock solution that was stored at −20°C. Samples of HD5 were prepared by diluting the stock solution to 5 µM in a total of 20 µL of 5 mM ammonium acetate pH 8 to give a final pH of 7.5. Fully reduced HD5 was prepared by adding 10 mM 2-mercaptoethanol to the ammonium acetate solution containing HD5 and incubating for >10 min at room temperature. Partially reduced HD5 was prepared by adding 2 mM 2-mercaptoethanol to the ammonium acetate solutions and incubating at room temperature for the indicated 10, 20, 40, or 60 min. For IMMS experiments, we added 1.5 µL of 2.5% acetic acid in MilliQ immediately prior to the measurements to lower the pH down to 5. This step was included because HD5 sprays much more efficiently under these conditions compared to pH 7.5. Both HD5ox and HD5red gave identical IMMS spectra (other than signal-to-noise) compared to the pH 5 data presented in the paper (data not shown). Acidification of the sample before analysis has the additional benefit of quenching the reduction reaction, as we found no evidence of either oxidation or further reduction up to 1 h after loading the samples for analysis (data not shown). For the tandem MS experiment, samples were alkylated and desalted prior to analysis. The reduction reaction was performed as described for the IMMS experiment. After reduction, a final concentration of 25 mM iodoacetamide was added to the solutions and incubated for 2 min at room temperature. Excess iodoacetamide was removed by desalting the samples with C18 ZipTips. The resin was washed twice with 12 µL of isopropanol followed by a wash with 12 µL of 0.1% acetic acid in MilliQ water. When the alkylation reaction was completed, the sample was pipetted over the resin repeatedly, which was then repeatedly washed in two separate volumes of 20 µL of 0.1% acetic acid and the sample eventually eluted in 4 µL of 80% isopropanol with 0.1% acetic acid in MilliQ water. The resulting samples were diluted 50-fold in 0.1% acetic acid in MilliQ water before direct infusion electrospray ionization.
IMMS
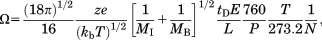 (1)
(1)Tandem MS
Samples of partially reduced/alkylated HD5 were analyzed on a Thermo Fisher Scientific Orbitrap Elite mass spectrometer. Samples were analyzed by direct infusion using a 100 µL glass syringe in a syringe pump set to 500 nL min−1. We used a New Objective Picotip Silicatip emitter with an inner tip diameter of 10 µm. The AGC targets were: full MS 1e6, SIM 1e5, MSn 2e5. S-lens RF was set to 60 V. The capillary temperature was set to 275°C. For CID spectra, we used a normalized collision energy of 35%. ETcaD spectra were acquired using a 20 ms reaction time, a reagent AGC target of 1e6 and 20% supplemental activation energy. The number of microscans was set to 5. All MS was performed with the Orbitrap analyzer, operated at a setting of 60,000 resolution. Both 6+ and 5+ precursors were selected for tandem MS CID and ETcaD. Spectra were processed using Protein Deconvolution 3.0, Manual Xtract. Peaks were matched using a modified version of SlinkS, using a 10 ppm mass window.37
Acknowledgments
The authors declare no conflict of interest. The authors thank Dr. Fan Liu for technical assistance in analyzing the tandem MS data with SlinkS.