Cerebrospinal fluid proteins in idiopathic intracranial hypertension: An exploratory SWATH proteomics analysis
Awadh Kishor Pandit and Shubham Misra have contributed equally this work and share the first authorship of the manuscript.
Abstract
Purpose
The pathogenesis of idiopathic intracranial hypertension (IIH) is currently poorly understood. This exploratory study aimed to identify potential cerebrospinal fluid (CSF) biomarkers in IIH cases compared to controls using SWATH-MS proteomics approach.
Experimental Design
CSF samples were collected prospectively from IIH cases and control subjects which were subjected to SWATH-MS based untargeted proteomics. Proteins with fold change > 1.5 or < 0.67 and p-value < 0.05 were considered significantly differentially expressed. Data are available via ProteomeXchange with identifier PXD027751. Statistical analysis was conducted in R version 3.6.2.
Results
We included CSF samples from 33 subjects, consisting of 13 IIH cases and 20 controls. A total of 262 proteins were identified in Proteinpilot search. Through SWATH analysis, we quantified 232 proteins. We observed 37 differentially expressed proteins between the two groups with 24 upregulated and 13 downregulated proteins. There were two differential proteins among overweight versus non-overweight IIH cases. Network for 23 proteins was highly connected in the interaction analysis.
Conclusions and Clinical Relevance
Neurosecretory, neuroendocrine, and inflammatory proteins were predominantly involved in causing IIH. This exploratory study served as a platform to identify 37 differentially expressed proteins in IIH and also showed significant differences between overweight and non-overweight IIH patients.
Abbreviations
-
- BMI
-
- body mass index
-
- CSF
-
- cerebrospinal fluid
-
- FDR
-
- false discovery rate
-
- GO
-
- gene ontology
-
- IIH
-
- idiopathic intracranial hypertension
-
- IQR
-
- interquartile range
-
- KEGG
-
- Kyoto encyclopedia of genes and genomes
-
- MRI
-
- magnetic resonance imaging
-
- S.D.
-
- standard deviation
-
- SWATH-MS
-
- sequential window acquisition of all theoretical fragment ion spectra- mass spectrometry
1 INTRODUCTION
Idiopathic Intracranial Hypertension (IIH) is a condition typically seen in women with raised intracranial pressure, without evidence of intracranial pathology [1]. There are studies detailing the risk factors associated with IIH but no evidence exists regarding these risk factors except for obesity [2]. There are various possible pathomechanism of the disease such as increased cerebrospinal fluid (CSF) secretion [3], increased CSF outflow resistance [4-6] or raised cerebral venous pressure [7] leading to imbalance of CSF and blood flow. The causes underlying the pathophysiology of IIH have been poorly understood. The CSF protein biomarkers of the disease may provide an insight into the pathomechanism of IIH and more relevance regarding vision impairment. The expected outcome of various candidate markers shall be of importance for therapeutic intervention in IIH and specifically early intervention to reduce vision impairment. Recent developments in proteomics approaches have shown the potential to discover CSF biomarkers in IIH. In a study published by Lecube et al. in 2012 [8], they performed quantitative proteomics in CSF comparing obese versus non-obese women with IIH. They proposed inflammatory, neuroendocrine, antioxidant, and somnogenic factors and brain plasticity-related proteins which may have a role in the pathogenesis of IIH in obese women. Therefore, using an untargeted proteomics approach, we conducted a pilot exploratory study to identify CSF proteins that may relate to the pathophysiology of disease and vision impairment in IIH. We further conducted a comparative analysis to identify the differentially expressed CSF proteins between overweight versus non-overweight IIH cases.
2 METHODS
2.1 Study population
This hospital-based case-control study was conducted in the Department of Neurology, All India Institute of Medical Sciences (AIIMS), New Delhi, India in collaboration with CSIR-Institute of Genomics and Integrative Biology (IGIB), New Delhi, India from 2018 to 2020. Patients aged between 18 and 50 years of either sex having IIH as per the modified Dandy Criteria [9] were recruited prospectively in the study. A total of 13 IIH patients were included in the study. Patients were excluded if pregnant, associated malignancy, chronic medical conditions, and not given consent to the study.
A control group comprising 20 individuals was recruited for the study. The control CSF were collected from non-head injury trauma cases and degenerative joint disease patients undergoing spinal anesthesia. Prior written informed consent was taken from all the subjects recruited in the study before CSF collection. The study was approved by the Local Institutional Ethics Committee of AIIMS, New Delhi (Ref. No.: IEC-371/06.07.2018).
2.2 Clinical evaluation
All the patients included in the study underwent detailed clinical examination which included demographic data, clinical symptoms, general medical examination, and detailed eye examination. Body mass index (BMI) more than 25 was considered overweight and was compared with the non-overweight subjects in the study. Eye examination included visual acuity and papilledema assessment. Additionally, other cranial nerve involvements and detailed neurologic examinations were also performed. Included patients had to undergo neuroimaging in the form of magnetic resonance imaging (MRI) brain along with MR venography.
2.3 CSF collection
The included patients underwent lumbar puncture for the CSF examination. Opening pressure was measured in all the patients and CSF was examined for total cells, glucose, protein, bacterial, fungal, and cytopathological examination to rule out an alternate diagnosis. The collected CSF was stored in −80°C with proper labeling until further analysis. No damage to the CSF specimen was assured during collection, cold chain maintenance, transportation, and storage.
2.4 Sample preparation
The proteins from CSF samples collected in the cryo-vials were diluted with 1× phosphate buffer saline (PBS) and precipitated in pre-chilled acetone overnight (12 h) at −20°C. The samples were then centrifuged at 15,000 × g for 15 min at 4°C and supernatants were discarded. The CSF protein pellet was resuspended in 0.1 M Tris-HCl with 8 M urea, pH 8.5.
2.5 Reduction, alkylation, and trypsin digestion
Twenty micrograms of protein from each sample was taken in fresh microcentrifuge tubes and diluted 10 times to bring urea concentration below 1 M before proceeding with reduction, alkylation, and trypsin digestion. 300 µg protein pooled from all the cases and controls were reduced with 25 mM of Dithiothreitol (DTT) for 30 min at 56°C, followed by alkylation using 55 mM of iodoacetamide (IAA) at room temperature (in the dark) for 20 min. These samples were then subjected to trypsin digestion in an enzyme-to-substrate ratio of 1:10 (trypsin: protein) for 16 h at 37°C. The tryptic peptides were vacuum dried in vacuum concentrator.
2.6 SWATH-based untargeted proteomics
2.6.1 Spectral ion library preparation
The tryptic peptides from pooled protein were fractionated into eight fractions using a strong cation exchange (SCX) cartridge (5 micron, 300 Å bead from SCIEX) and an increasing concentration of ammonium formate buffer (35, 50, 75, 100, 125, 150, 250, and 350 mM ammonium formate, 30% v/v acetonitrile; pH = 2.9). Each eluted fraction was desalted using a C18 Ziptip (Merck, USA) and analyzed on a quadrupole-TOF hybrid mass spectrometer (TripleTOF 6600, SCIEX) coupled to an Eksigent NanoLC-425 system in data-dependent acquisition (DDA) mode. Peptides were loaded on a trap-column (ChromXP C18CL 5 µm 120 Å, Eksigent, SCIEX) and online desalting was performed with a flow rate of 10 µL/min for 10 min. Peptides were separated on a reverse-phase C18 analytical column (ChromXP C18, 3 µm 120 Å, Eksigent, SCIEX) in a 57-min-long gradient with a flow rate of 5 µL/min using buffer A (water with 0.1% formic acid) and buffer B (acetonitrile with 0.1% formic acid).
Optimized source parameters were used, curtain gas and nebulizer gas were maintained at 25 and 30 psi, respectively, the ion spray voltage was set to 5.5 kV and the temperature was set to 250°C. For DDA, a 1.8-s instrument cycle was repeated in high sensitivity mode throughout the whole gradient, a full scan MS spectrum for 400–1250 m/z was performed with an accumulation time of 250 ms, followed by 30 MS/MS experiments with 50 ms accumulation time and 100–1500 m/z range each, on MS precursors with charge state 2 + to 5 + exceeding a 120-cps threshold. The rolling collision energy was used and former target ions were excluded for 15 s.
Statement of Clinical Relevance
The pathogenesis of idiopathic intracranial hypertension (IIH) is currently poorly understood. Using an untargeted proteomics approach, our study aimed to identify all the CSF proteins that may relate to the pathophysiology of disease and vision impairment in IIH. This is the first study which identified the potential protein biomarkers in IIH cases compared to control subjects using the SWATH-MS approach. We also determined differentially expressed proteins in CSF between overweight and non-overweight IIH cases. Our exploratory study provided a list of potential differentially expressed protein biomarkers in IIH cases which may relate to the pathophysiology of the disease. We emphasized that CSF is a definite fluid that could represent the potential protein biomarkers involved in causing IIH. The results of this exploratory study could serve as a platform to conduct future targeted proteomics studies on a large sample to better elucidate the role of these candidate proteins in the pathomechanism of IIH.
2.6.2 SWATH data acquisition
We performed proteomics analysis using CSF without depletion of abundant proteins. SWATH-MS analysis for the samples was performed on the same instrument setup, chromatographic conditions, and source parameters as DDA. Four micrograms of desalted peptides were analyzed from each sample in a SWATH method with 100 precursor isolation windows, defined based on precursor m/z frequencies in a DDA run using the SWATH Variable Window Calculator (SCIEX), with a minimum window of 5 m/z. Data were acquired using Analyst TF 1.7.1 Software (SCIEX). Accumulation time was set to 250 ms for the MS scan (400–1250 m/z) and 25 ms for the MS/MS scans (100–1500 m/z). Rolling collision energies were applied for each window based on the m/z range of each SWATH and a charge 2+ ion, with a collision energy spread of 5. The total cycle time was 2.7 s.
2.7 Bioinformatics and statistical analyses
A combined database search was performed for .wiff format raw files generated in DDA mode against UniProtKB human FASTA database (Swissprot and TrEMBL; 152,774 entries) using ProteinPilot Software 5.0.1 (SCIEX). The group output file from ProteinPilot served as the spectral ion library for SWATH analysis. Protein identification in ProteinPilot software was performed using the default settings. Cysteine alkylation through iodoacetamide was set as fixed modification, maximum two missed cleavages were allowed, biological modification was activated in the “ID focus,” search effort was set to “Thorough ID,” detected protein threshold [Unused ProtScore (conf)] was set to 0.05 (10.0%) and false discovery analysis was activated. A 1% global false discovery rate (FDR) at the protein level, excluding shared peptides was considered for import in SWATH 2.0 microapp of PeakView 2.2 software (SCIEX). SWATH run files were added and retention time alignment was performed using peptides from abundant proteins. For SWATH analysis, sequence unique peptides were used for quantification. The processing settings for peak extraction were: maximum of 10 peptides per protein, 5 transitions per peptide, > 95% peptide confidence threshold, 1% peptide FDR. XIC extraction window was set to 55 min, with 75 ppm XIC Width. The protein area was exported in the form of MarkerView (.mrkw) file. In MarkerView 1.2.1 (SCIEX), data normalization was performed using total area sum normalization. There were no missing values in the dataset.
The data were Log2 transformed as significant skewness or kurtosis was present in the peak area values. After Log2 transformation and confirmation of normality in the data distribution using the Shapiro-Wilk test, continuous variables were represented by mean and standard deviation (SD) or median and interquartile range (IQR) and were analyzed using student the t-test. Dichotomous variables were represented by number, percentage and were analyzed using the Chi-square test. Differentially expressed proteins were identified using a Log2 fold change cut-off of > 0.58 (corresponding to fold change > 1.5) for upregulation and < −0.58 (corresponding to < 0.67-fold change) for downregulation and a Log10 p-value cut-off of > 1.3 (corresponding to p-value < 0.05). Receiver operating characteristic (ROC) curves were developed and area under the curve (AUC) was calculated for differentially expressed proteins. Protein interaction network analysis of significantly differentially expressed proteins was performed using STRING (version 11; http://string-db.org/) [10] and Cytoscape 3.9.0 software [11]. Functional analysis was performed using g:Profiler (https://biit.cs.ut.ee/gprofiler/gost) [12]. The results were reported in accordance with the STROBE guidelines [13]. Statistical analysis was conducted in R version 3.6.2 using the following packages: “plyr”, “tidyverse”, “gplots”, and “pheatmap”.
3 Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium [14] via the PRIDE [15] partner repository with the dataset identifier PXD027751.
4 RESULTS
4.1 Baseline characteristics of IIH cases and controls
Our study included CSF samples from 33 subjects, consisting of 13 IIH cases and 20 control subjects. The mean age of cases was 27.69 ± 6.94 and controls was 37.35 ± 13.72 (p = 0.03). There were 11 (84.61%) females in cases and six (30%) in controls (p = 0.02). The duration of symptoms ranged from 2 to 28 weeks. The most common symptoms were headache and transient visual obscurations in the IIH group. The visual acuity and neuroimaging findings are given in Table 1. The details of controls are described in Table 2. All the patients recruited as controls in our study were having trauma or any orthopedics surgery and had undergone spinal anesthesia during which CSF was collected for the study after a written valid consent.
| S. No. | Age (years) | Sex | Duration of symptoms (weeks) | Symptoms | Cranial nerves involved | Visual acuity | Papilledema grade | MRI brain | MR venography | CSF Pressure (mm of CSF) | Treatment | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Right | Left | |||||||||||
| 1 | 21 | M | 24 | Headache | II | 6/18 | 6/18 | Grade 1 | Normal | Normal | 150 | Acetazolamide |
| 2 | 33 | F | 28 | Headache, TVOs | II, VI | 6/18 | 6/12 | Grade V | Normal | Normal | 150 | Acetazolamide |
| 3 | 35 | F | 04 | Headache, TVOs | II, VI | 6/36 | PL+ | Grade III | Abnormala | Normal | 25 | Acetazolamide |
| 4 | 22 | M | 06 | Headache | II | 6/24 | 6/60 | Grade IV | Normal | Normal | 15 | Acetazolamide |
| 5 | 22 | F | 02 | Headache, TVOs | II | 6/6 | 6/6 | Grade IV | Normal | Normal | 39 | Acetazolamide |
| 6 | 38 | F | 20 | Headache, TVOs | II | 6/6 | 6/6 | Grade III | Normal | Normal | 40 | Acetazolamide |
| 7 | 34 | F | 24 | Headache, TVOs | II | 6/18 | 6/18 | Grade IV | Abnormala | Normal | 160 | Acetazolamide |
| 8 | 23 | F | 04 | Headache, TVOs | II | 3/60 | 3/24 | RE Grade II and LE Grade I | Normal | Abnormalb | 400 | Acetazolamide, Topiramate |
| 9 | 18 | F | 18 | Headache, TVOs, Diplopia | II, VI | 6/18 | 6/12 | Grade V | Abnormala | Abnormalb | 380 | Acetazolamide, Topiramate |
| 10 | 33 | F | 06 | Headache | II | 6/18 | 6/24 | Grade III | Normal | Normal | 220 | Acetazolamide |
| 11 | 23 | F | 23 | Headache, Diplopia | II | PL+ | PL+ | Grade IV | Abnormala | Abnormalb | 22 | Acetazolamide, Topiramate |
| 12 | 35 | F | 04 | Headache, TVOs | II | 6/18 | 6/9 | Grade III | Abnormala | Normal | 350 | Acetazolamide |
| 13 | 23 | F | 20 | Headache, TVOs | II, VI | 6/24 | 6/12 | Grade V | Normal | Normal | 160 | Acetazolamide |
- Abbreviations: F, female; LE, left eye; M, male; PL, perception of light; RE, right eye; TVOs, transient visual obscurations.
- a Empty sella, scleral flattening, optic nerve protrusion, optic nerve sheath distention, optic nerve tortuosity.
- b Left transverse sinus stenosis.
| S. No. | Age (years) | Sex | Diagnosis |
|---|---|---|---|
| 1 | 55 | Female | Bilateral knee osteoarthritis (OA) with left knee replacement |
| 2 | 23 | Male | Fracture right fibula |
| 3 | 64 | Female | Left knee OA for left knee replacement |
| 4 | 51 | Female | Left hip OA for left hip replacement |
| 5 | 51 | Male | Right knee OA for right knee replacement |
| 6 | 24 | Male | Multiple fracture (shaft of femur and humerus) for open reduction and internal fixation |
| 7 | 49 | Male | Left intertrochanteric fracture for closed reduction and internal fixation |
| 8 | 20 | Male | Road traffic accident for right leg debridement |
| 9 | 22 | Female | Degloving injury right lower limb surgery |
| 10 | 17 | Female | Shaft of femur fracture for surgery—implant removal |
| 11 | 33 | Male | Left both bone crush Injury |
| 12 | 40 | Male | Trauma without head injury |
| 13 | 35 | Male | Trauma without head injury |
| 14 | 26 | Male | Trauma without head injury |
| 15 | 49 | Male | Trauma without head injury |
| 16 | 32 | Male | Trauma without head injury |
| 17 | 51 | Male | Trauma without head injury |
| 18 | 24 | Male | Trauma without head injury |
| 19 | 42 | Male | Trauma without head injury |
| 20 | 39 | Female | Trauma without head injury |
4.2 Identification of differentially expressed proteins in IIH cases and controls using SWATH-MS/MS
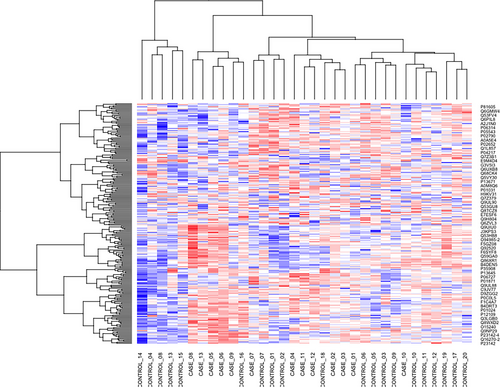
Total 262 protein groups were identified in ProteinPilot search in 1% global FDR. Through SWATH analysis, we could quantify 232 proteins in the two groups of samples. A total of 37 differentially expressed proteins were identified between the two groups (Figure 1). Out of 37 proteins, 24 proteins were upregulated while 13 proteins were downregulated in IIH cases compared to the control subjects (Table 3). Nineteen out of 37 proteins were upregulated/downregulated by more than two-folds. Complement C1q subcomponent subunit B was found to be six-fold upregulated in IIH cases while Myosin-reactive immunoglobulin kappa chain variable region was downregulated by 11-fold. A heatmap of all the 232 identified proteins is given in Figure 2. The AUC of 37 differentially expressed proteins is given in Table S1.
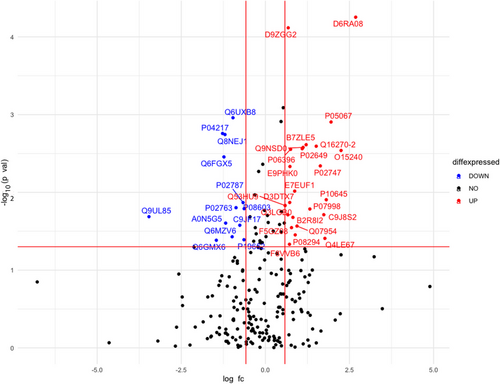
| S. No | Protein ID | Protein name | Mean case | Mean control | Log2 fold change | Fold change | Log10 p-value | p-value |
|---|---|---|---|---|---|---|---|---|
| 1 | D6RA08 | Complement C1q subcomponent subunit B (Fragment) | 15.26 | 12.58 | 2.68 | 6.42 | 4.25 | <0.001 |
| 2 | O15240 | Neurosecretory protein VGF | 18.53 | 16.28 | 2.24 | 4.75 | 2.53 | 0.002 |
| 3 | P05067 | Amyloid beta A4 protein | 20.89 | 18.93 | 1.95 | 3.87 | 2.90 | 0.001 |
| 4 | P10645 | Chromogranin-A | 20.60 | 18.79 | 1.81 | 3.50 | 1.90 | 0.01 |
| 5 | Q4LE67 | CSPG3 variant protein (Fragment) | 18.04 | 16.27 | 1.76 | 3.40 | 1.40 | 0.03 |
| 6 | C9J8S2 | Retinoic acid receptor responder protein 2 (Fragment) | 16.09 | 14.35 | 1.73 | 3.33 | 1.71 | 0.01 |
| 7 | P02747 | Complement C1q subcomponent subunit C | 20.21 | 18.57 | 1.63 | 3.10 | 2.34 | 0.004 |
| 8 | Q16270-2 | Isoform 2 of Insulin-like growth factor-binding protein 7 | 21.00 | 19.49 | 1.51 | 2.85 | 2.59 | 0.002 |
| 9 | P07998 | Ribonuclease pancreatic | 19.13 | 17.80 | 1.32 | 2.50 | 1.78 | 0.01 |
| 10 | B7ZLE5 | FN1 protein | 20.96 | 19.75 | 1.21 | 2.31 | 2.61 | 0.002 |
| 11 | Q9NSD0 | Protein S | 19.35 | 18.22 | 1.12 | 2.17 | 2.57 | 0.002 |
| 12 | P02649 | Apolipoprotein E | 24.70 | 23.61 | 1.09 | 2.13 | 2.56 | 0.002 |
| 13 | Q07954 | Prolow-density lipoprotein receptor-related protein 1 | 12.73 | 11.78 | 0.94 | 1.91 | 1.56 | 0.02 |
| 14 | P08294 | Extracellular superoxide dismutase [Cu-Zn] | 21.62 | 20.73 | 0.88 | 1.84 | 1.45 | 0.03 |
| 15 | E7EUF1 | Ectonucleotide pyrophosphatase/phosphodiesterase family member 2 | 22.23 | 21.35 | 0.870 | 1.82 | 2.01 | 0.009 |
| 16 | B2R8I2 | cDNA, FLJ93914, highly similar to Homo sapiens histidine-rich glycoprotein (HRG), mRNA | 19.99 | 19.17 | 0.81 | 1.76 | 1.67 | 0.02 |
| 17 | F5GZ08 | Amyloid-like protein 1 | 20.24 | 19.46 | 0.77 | 1.71 | 1.54 | 0.02 |
| 18 | P06396 | Gelsolin | 21.89 | 21.14 | 0.74 | 1.68 | 2.55 | 0.002 |
| 19 | E9PHK0 | Tetranectin | 17.64 | 16.91 | 0.73 | 1.65 | 2.33 | 0.004 |
| 20 | D3DTX7 | Collagen, type I, alpha 1, isoform CRA_a | 15.45 | 14.72 | 0.72 | 1.65 | 1.87 | 0.01 |
| 21 | F8VVB6 | Protein kinase C-binding protein NELL2 | 19.69 | 18.97 | 0.71 | 1.64 | 1.33 | 0.04 |
| 22 | D9ZGG2 | Vitronectin | 21.95 | 21.27 | 0.67 | 1.60 | 4.11 | <0.001 |
| 23 | Q3LGB0 | Osteopontin (Fragment) | 19.86 | 19.19 | 0.66 | 1.58 | 1.71 | 0.01 |
| 24 | Q53HU9 | Complement component 1, r subcomponent variant (Fragment) | 18.77 | 18.18 | 0.58 | 1.50 | 1.83 | 0.01 |
| 25 | Q9UL85 | Myosin-reactive immunoglobulin kappa chain variable region (Fragment) | 16.23 | 19.69 | -3.46 | 0.09 | 1.68 | 0.02 |
| 26 | Q6GMX6 | IGH@ protein | 15.97 | 17.42 | -1.45 | 0.36 | 1.38 | 0.04 |
| 27 | P04217 | Alpha-1B-glycoprotein | 21.68 | 22.96 | -1.27 | 0.41 | 2.75 | 0.001 |
| 28 | Q6FGX5 | TIMP metallopeptidase inhibitor 1, isoform CRA_a | 17.47 | 18.70 | -1.23 | 0.42 | 2.45 | 0.003 |
| 29 | Q8NEJ1 | Uncharacterized protein | 19.33 | 20.53 | -1.19 | 0.43 | 2.74 | 0.001 |
| 30 | A0N5G5 | Rheumatoid factor D5 light chain (Fragment) | 19.35 | 20.54 | -1.18 | 0.44 | 1.60 | 0.02 |
| 31 | Q6MZV6 | Putative uncharacterized protein | 17.51 | 18.50 | -0.98 | 0.50 | 1.42 | 0.03 |
| 32 | Q6UXB8 | Peptidase inhibitor 16 | 17.06 | 18.02 | -0.96 | 0.51 | 2.95 | 0.001 |
| 33 | P02763 | Alpha-1-acid glycoprotein 1 | 24.41 | 25.28 | -0.87 | 0.54 | 1.80 | 0.01 |
| 34 | C9JF17 | Apolipoprotein D (Fragment) | 22.66 | 23.42 | -0.76 | 0.59 | 1.57 | 0.02 |
| 35 | P02787 | Serotransferrin | 22.07 | 22.73 | -0.66 | 0.63 | 1.86 | 0.01 |
| 36 | P19652 | Alpha-1-acid glycoprotein 2 | 20.80 | 21.43 | -0.63 | 0.64 | 1.39 | 0.04 |
| 37 | P08603 | Complement factor H | 19.76 | 20.39 | -0.63 | 0.64 | 1.79 | 0.01 |
- Note: The data was log2 transformed therefore, log2 fold change was calculated using the formula: log2FC = log2 (mean case)- log2 (mean control). Absolute ratio for mean case/ mean control was the anti-log2 of the log2 fold change value.
A subgroup analysis was conducted to identify the differentially expressed proteins between overweight and non-overweight IIH cases. We observed only two proteins (Oligodendrocyte myelin glycoprotein variant and Semaphorin-7A) both upregulated in nine overweight IIH cases compared to four non-overweight IIH cases (Figure S1 and Table S2).
4.3 Protein-protein interactions
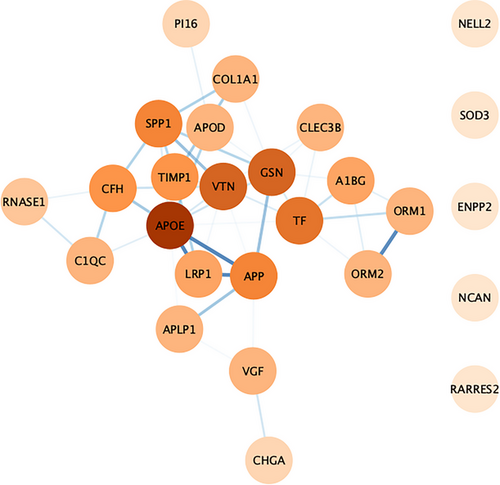
The 37 differentially expressed proteins were loaded into the STRING online tool (Search Tool for the Retrieval of Interacting Genes/Proteins 11). The network nodes represent proteins while the edges represent the protein-protein interactions. Out of 37 proteins, 26 were successfully matched to proteins within the STRING database, while 11 were unmatched due to insufficient interaction data. This interaction network was further analyzed using the Cytoscape 3.9.0 software. The interaction network consisted of 26 nodes and 46 edges. The interaction network for 21 proteins was found to be highly connected showing molecular interactions with other proteins while five proteins (NELL2, SOD3, ENPP2, NCAN, and RARRES2) were either not connected or had interactions of low confidence with the entire network. APOE had a strong interaction profile with other proteins with the highest degree of interaction = 11 followed by VTN and GSN having a degree of interaction = 8. LRP1-APOE had the highest protein-protein interaction score of 0.999 followed by APOE-APP (score = 0.998), LRP1-APP (score = 0.992), and ORM1-ORM2 (score = 0.984). The interaction network is depicted in Figure 3.
4.4 Functional analysis using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG)
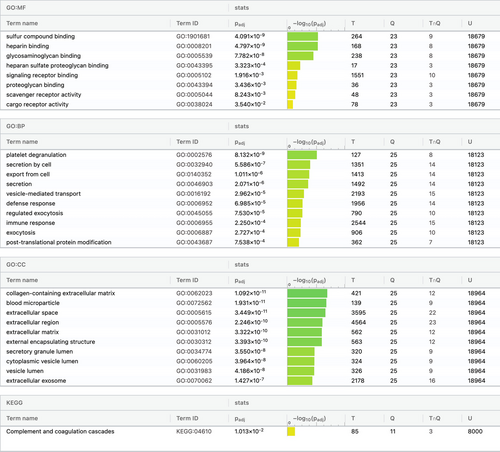
The functional analysis was performed using the g:Profiler online tool. The top 10 highly significant pathways/processes were selected. In the GO enrichment analysis, the most significant molecular functions included sulfur compound binding, heparin binding and glycosaminoglycan binding. Platelet degranulation, secretion by cell, and export from cell were found to be the most significant biological processes, while the cellular components included collagen-containing extracellular matrix, blood microparticles, and extracellular space. The KEGG pathway enrichment analysis observed that the significantly differentially expressed proteins were involved in the complement and coagulation cascade pathway (Figure 4).
4.5 Biological roles of significantly differentially expressed proteins
The proteins that were significantly differentially expressed in IIH cases compared to control subjects were categorized into various biological functions including neuroendocrine and neurosecretion, inflammation, antioxidants, adhesion molecules/apoptosis, and proteins involved in cell proliferation (Table S3).
5 DISCUSSION
Our pilot exploratory study identified 37 potential protein biomarkers differentially expressed in IIH cases compared to control subjects using a discovery-based untargeted proteomics approach. Out of 37 proteins, 26 might have a potential role in the pathogenesis of IIH. We observed that neurosecretory, neuroendocrine, and inflammatory proteins were predominantly involved in causing IIH, among all the significant proteins. The biological processes implicating the pathomechanism of IIH were: increased CSF secretion, inflammation, and coagulation cascade. The various proteins leading to these biological processes are as described in Table S1. We emphasize that CSF is a definite fluid that could represent the potential protein biomarkers involved in causing IIH.
To the best of our knowledge, there has been no such study to date that has explored proteins in cases of IIH using a discovery-based SWATH-MS approach. However, a study published by Lecube et al. in 2012 [8] did a comparative analysis of obese versus non-obese proteins in 16 IIH cases and observed 18 differentially expressed proteins using a quantitative LC-MS proteomics approach. The differential production of only four (Osteopontin, Neurosecretory protein VGF, Chromogranin-A, and Fibrinogen- γ) out of 18 proteins was confirmed upon validation using Enzyme Linked Immunosorbent Assay (ELISA) or western blotting. Comparing their results with our study, five out of 37 significant proteins (Neurosecretory protein VGF, Chromogranin-A, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, Osteopontin, and Alpha-1-acid glycoprotein 2) were common and were also differentially expressed in the study by Lecube et al. 2012. However, when we analyzed the results according to the BMI data in our study, we observed only Oligodendrocyte myelin glycoprotein variant and Semaphorin-7A upregulated in overweight IIH cases; both of which were not previously identified by Lecube et al. 2012 [8]. Further validation of differentially expressed proteins identified in obese/overweight IIH cases in our study and Lecube et al. 2012 [8] study is required in a larger cohort of patients using targeted proteomics approach.
The role of CSF proteins as potential biomarkers in understanding the pathophysiology of IIH has been investigated in a few studies using immunoassays or two-dimensional fluorescence differential in-gel electrophoresis (2-D DIGE). A recent study published by Beier et al. in 2020 reported CSF neurofilament having significant association with permanent optic nerve damage in 61 cases of IIH [16]. Samanci et al. in 2017 studied the role of adipokines and cytokines as prognostic biomarkers for IIH to predict relapse and observed that the serum levels of IL-1β were significantly higher and of IL-8 and TNF- α significantly lower in 36 IIH cases compared to 40 healthy controls [17]. Doppler et al. in 2016 observed that the levels of aquaporin-4 were decreased in CSF, and leptin was increased in CSF and plasma of 28 IIH cases compared to 29 control subjects [18]. Altıokka-Uzun et al. in 2015 observed that patients with IIH had highly elevated cytokine levels including TNF-α, IFN-γ, IL-4, IL-10, IL-12, IL-17 in serum compared to patients with multiple sclerosis and healthy controls [19]. Brettschneider et al. in 2011 used 2-D DIGE and found six proteins that were upregulated in 18 IIH cases (sterol regulatory element-binding protein 1, zinc-alpha-2-glycoprotein, immunoglobulin heavy constant alpha 1 [IGHA1], alpha-1-antitrypsin [SERPINA1], serotransferrin, haptoglobin), and four proteins that were downregulated (hemopexin, angiotensinogen, vitamin-D-binding protein, transthyretin) compared to 18 control subjects [20]. This study further validated and showed the downregulation of angiotensinogen in patients with IIH and proposed the possibility of this candidate protein in increasing blood flow in the choroid plexus and further increased CSF production. However, in our study serotransferrin was found to be downregulated in IIH cases in contrast to the findings published by Brettschneider et al. in 2011 [20]. The difference in the expression pattern might result from the different types of control subjects (non-head injury trauma patients and degenerative joint diseases in our study and tension-type headache patients in Brettschneider et al.) included in the two studies. Further, the proteomics approach (SWATH-MS) used in our study is much more reliable and sensitive than 2-D DIGE used by Brettschneider et al. as SWATH-MS includes 10 peptides for each protein and 5 transitions for each peptide identified at 1% FDR at both protein and peptide level. However, validation studies might be needed to confirm these contrasting findings in the future. A study conducted by Sinclair et al. in 2010 showed in vitro choroid plexus (CP) and arachnoid granulation tissue (AGT) and follow-up in vivo CP and AGT found; 11beta-hydroxysteroid dehydrogenase type 1 (11beta-HSD1) and key elements of the glucocorticoid signaling pathway were expressed in CP and AGT [21]. After weight loss (14.2 ± 7.8 kg; p < 0.001), global 11-HSD1 activity decreased (p = 0.001) and correlated with reduction in intracranial pressure (r = 0.504; p = 0.028). CSF and serum glucocorticoids remained stable, although the change in CSF cortisone levels correlated with weight loss (r = −0.512; p = 0.018). The role of inflammation in the pathophysiology of IIH was further confirmed by Edwards et al. in 2010 which concluded that IL-17 is more likely to be detected in the CSF of IIH patients compared to chronic inflammatory demyelinating peripheral neuropathy (CIDP), clinically isolated syndrome (CIS), and multiple sclerosis [22].
Female gender of childbearing age is one of the most common risk factors of IIH [23]. Previous studies have demonstrated higher levels of 5 alpha-reductase and 11 beta- hydroxysteroid dehydrogenase 1 (HSD1) in females both of which are regulated by estrogen, but their role in IIH patients warrants further investigation [24, 25]. In our study, 17 proteins were differentially expressed between males and females, of which 13 were upregulated in females (Figure S2 and Table S4). However, due to the imbalance in the two groups, these findings are inconclusive. Future studies with large sample sizes are required to explain the pathomechanism behind the female gender being a risk factor for IIH.
However, a limitation of our study is the lack of absolute quantification data for the identified proteins. Our pilot study identified potential protein biomarkers and lacked a validation cohort. The prevalence of females was lower in the control group. Thus, our findings must be interpreted as exploratory and the studies in future must validate our results in a large sample of IIH cases and controls using different targeted proteomics or immunoassay approaches.
6 CONCLUSION
To summarize, our study identified 37 differentially expressed proteins in the CSF of IIH cases using SWATH-MS proteomics analysis. Several of these proteins were associated with biological functions including neuroendocrine and neurosecretion, inflammation, antioxidants, adhesion molecules/apoptosis and cell proliferation, thereby providing an insight into the possible pathomechanism of IIH. We further identified proteins in overweight IIH cases and proteins differentially expressed in females compared to males. Our exploratory study, in the future, should serve as a platform to validate these potential candidate biomarkers in a large set of IIH patients to elucidate the etiology and pathomechanism of IIH better.
ACKNOWLEDGMENTS
A.K. Pandit received funding from the AIIMS Intramural Grant to support this study. S. Misra is a DST-INSPIRE Fellow supported by Department of Science and Technology, Government of India. B. Jee is a Scientist-C at Department of Health Research, Government of India. This work was supported by the AIIMS Intramural Research Grant (Grant number: F.8-649/A-649/2018/RS).
CONFLICT OF INTEREST STATEMENT
The authors declared no conflicts of interest.
Open Research
DATA AVAILABILITY STATEMENT
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD027751.