Viruses and Mycoplasma pneumoniae are the main etiological agents of community-acquired pneumonia in hospitalized pediatric patients in Spain
Enrique Otheo and Mario Rodríguez are both first authors.
Abstract
Objectives
To describe the etiology of community-acquired pneumonia (CAP) in hospitalized children in Spain and analyze the predictors of the etiology.
Hypothesis
The different etiological groups of pediatric CAP are associated with different clinical, radiographic, and analytical data.
Design
Observational, multicenter, and prospective study.
Patient selection
This study included children aged 1 month to 17 years with CAP, who were hospitalized between April 2012 and May 2019.
Methods
An extensive microbiological workup was performed. The clinical, radiographic, and analytical parameters were analyzed for three etiological groups.
Results
Among the 495 children included, at least one causative pathogen was identified in 262 (52.9%): pathogenic viruses in 155/262 (59.2%); atypical bacteria (AB), mainly Mycoplasma pneumonia, in 84/262 (32.1%); and typical bacteria (TyB) in 40/262 (15.3%). Consolidation was observed in 89/138 (64.5%) patients with viral CAP, 74/84 (88.1%) with CAP caused by AB, and 40/40 (100%) with CAP caused by TyB. Para-pneumonic pleural effusion (PPE) was observed in 112/495 (22.6%) patients, of which 61/112 (54.5%) presented a likely causative pathogen: viruses in 12/61 (19.7%); AB in 23/61 (37.7%); and TyB in 26/61 (42.6%). Viral etiology was significantly frequent in young patients and in those with low oxygen saturation, wheezing, no consolidation, and high lymphocyte counts. CAP patients with AB as the etiological agent had a significantly longer and less serious course as compared to those with other causative pathogens.
Conclusions
Viruses and M. pneumoniae are the main causes of pediatric CAP in Spain. Wheezing, young age, and no consolidation on radiographs are indicative of viral etiology. Viruses and AB can also cause PPE. Since only a few cases can be directly attributed to TyB, the indications for antibiotics must be carefully considered in each patient.
1 INTRODUCTION
Community-acquired pneumonia (CAP) is a major cause of hospitalization and morbidity in children.1 Annually, approximately 20 million children with CAP need hospitalization worldwide.2
Important studies have been performed in recent years on the etiology of CAP, and the knowledge of relevant etiological data is critical for adequate policies and improved management of CAP. Conventional microbiological studies can identify the etiology in only a limited number of patients. Sampling the lower respiratory tract in children is challenging, and contamination with colonizing organisms is frequent in samples obtained from the upper respiratory tract.3 Direct sampling from the lung is impractical and rarely accepted, and bronchoalveolar lavage samples only yield positive results in 29% of cases.4 Direct sampling is considered a standard of care only in children with large para-pneumonic effusion (PPE), which is the most common complication of CAP in this population group. Several institutions, including the World Health Organization (WHO), are encouraging improvements in the diagnostic tests for CAP.5
Recently, with the arrival of new microbiology techniques based on molecular biology, viruses have emerged as the most common etiological agents of CAP. Unfortunately, these techniques are not available at all centers for all patients, and most pediatric patients with CAP are administered antibiotics, regardless of hospital admission. In Spain, the data regarding the etiology of pediatric CAP are limited.6-8 Better knowledge of this condition is essential to educate clinicians, avoid the unnecessary use of antibiotics, contain the emergence of microbial resistance, and limit the impact of the disease on the child, family, and healthcare systems.9
This study aimed to describe the etiology of a cohort of hospitalized children and adolescents with CAP in Spain, including those with associated PPE. Additionally, we aimed to determine the clinical, analytical, and radiographic predictors of the etiology.
2 METHODS
2.1 Study design
This observational, multicenter, prospective cohort study was conducted in two phases. The first pilot phase was performed at two centers in Madrid, Spain from April 2012 to March 2015. Enrolment was expanded in the second phase to 15 centers in three regions of Spain (Madrid, Basque Country, and Andalusia) from December 2017 to May 2019. Both phases were approved by the Ethics Boards of the participating hospitals. The necessary information regarding the study was explained to the guardians and written consent was obtained. Similarly, information was provided to patients aged 12–17 years comprehensively, and their assent was obtained.
2.2 Collection of data
Relevant information for this study was encoded and entered into an electronic database. Confidentiality was maintained through the codification of patients. The basal features including clinical data, findings of imaging studies, biomarker test results, and microbiological findings were recorded. Two principal researchers, data managers, and biostatisticians exclusively handled the database.
2.3 Participants
The participants eligible for this study were hospitalized children and adolescents, aged 1 month to 17 years, with clinically suspected pneumonia as determined by the attending physician and pathologic findings on chest radiographs (CXR). Exclusion criteria were immunosuppressive conditions, chronic cardiac or pulmonary disease (except asthma), admission to a hospital in the last 30 days, and suspicion of lung aspiration or foreign body in the airway. The criteria for hospital admission included infants aged <6 months, poor feeding, dehydration, electrolyte disturbance, respiratory or hemodynamic instability, transcutaneous oxygen saturation (tcSatO2) ≤92% with fraction of inspired oxygen of 0.21, lethargy, apnea, pulmonary complication (PPE, necrosis, or abscess), multifocal involvement on CXR, failure to respond to oral antimicrobials after 48 h, and poor treatment compliance.
2.4 Procedures
On admission, a blood sample was collected for full blood cell count, standard biochemistry, and determination of inflammatory markers. Extensive microbiological workup was performed. Blood samples were obtained for cultures before starting antibiotics using the BacT/ALERT™ 3D blood culture system (Biomérieux) or BacTec™ 9240 blood culture system (Becton, Dickinson and Company). Culture with Streptococcus pneumoniae antigen in pleural fluid (PF; Alere BinaxNow™, S. pneumoniae antigen) was additionally performed in case of thoracentesis. Polymerase chain reaction (PCR) test was used to detect infectious agents in blood and nasopharyngeal aspirate (NPA) samples. The genetic material (DNA and RNA) was extracted for this procedure using an automated system (Nuclisens EasyMag bioMérieux), and the elutes were stored at −80°C until processing. The presence of S. pneumoniae in the blood samples was determined based on the detection of lytA genes, according to a previous report.10 In the NPA samples, the strategy of multiplex PCR for detecting up to 16 viruses, as well as Mycoplasma pneumoniae and Chlamydophila pneumoniae, was considered the best option. The respiratory viruses included respiratory syncytial virus (RSV) A and RSV-B; human metapneumovirus (hMPV); parainfluenza virus (PIV) 1, 2, 3, and 4; influenza virus (IV) A and B; human bocavirus (hBoV); adenovirus (ADV); enterovirus (EV); human rhinovirus (hRV); and human coronavirus (hCoV) 229E, OC43, NL63, and HKU12. Two paired samples for serology (at admission and after 2–4 weeks) of M. pneumoniae and C. pneumoniae were analyzed using a chemiluminescence assay (Virclia Monotest, Vircell). Only specimens obtained within 72 h after admission were included, except for PF samples, which were included if collected within 7 days after admission, and the second sample for serology.
- a)
Likely typical bacterial (TyB) infection: A bacterial pathogen (S. pneumoniae, Staphylococcus aureus, Streptococcus pyogenes, Haemophilus influenzae, etc.) detected in the blood through culture or PCR or in the PF through culture test or S. pneumoniae antigen detection; Staphylococcus epidermidis and other pathogens, typically considered contaminants in healthy children, were excluded.
- b)
Likely atypical bacterial (AB) infection: M. pneumoniae or C. pneumoniae detected by PCR in NPA or seroconversion or a significant increase in immunoglobulin G titers in the second sample, according to the manufacturer's recommendations.
- c)
Likely viral infection: At least one putative pathogen respiratory virus (RSV, PIV, hMPV, and IV) detected in NPA by PCR; other respiratory viruses (hRV, ADV, EV, hCoV, and hBoV) were not included as likely viral infections because of the poor likelihood that their detection in the upper respiratory tract indicates a pathogenetic role in lung infection.4, 11-14
The CXRs were interpreted according to the standards of the “WHO Vaccine Trial Investigators Radiology Working Group.”15 These standards establish two possible interpretations: primary end-point pneumonia (presence of consolidation, infiltrate, or pleural effusion), henceforth referred to as “consolidation,” and “other infiltrates.” PPE was diagnosed using CXR and confirmed by ultrasonography. This study applied the definitions by Hamm and Light regarding the types of PPE, that is, uncomplicated para-pneumonic effusions (ucPPE) and complicated para-pneumonic effusions (cPPE)/empyema.16
2.5 Statistical analysis
Baseline data regarding the sociodemographic, clinical, CXR, and analytical variables were described. Continuous variables are presented as medians and interquartile ranges (IQR) for nonparametric variables and means with standard deviations for normally distributed variables, previously tested with the Shapiro–Wilk and Kolmogorov–Smirnov tests. Categorical variables are presented as counts and percentages. The chi-squared or Fisher tests were applied for comparing the categorical variables and the Kruskal–Wallis or t-tests for comparing continuous variables. To determine the factors associated with different CAP etiologies, we performed a univariable multinomial regression. All hypothesis testing was performed at a 5% significance level. Plots and analyses were performed using the R software (R Foundation for Statistical Computing, Vienna).17
3 RESULTS
In total, 495 children were enrolled: 151 in phase 1 and 344 in phase 2. The demographic, clinical, analytical, and CXR characteristics are shown in Table 1. The number and proportions of the different microbiological tests performed are described in Table 2.
| N = 495 | |
|---|---|
| Categorical variables | N (%) |
| Male gender | 271 (54.7) |
| Antecedent of asthma or airway reactive disease | 94 (19) |
| Antibiotic before admission | 93 (18.8) |
| Wheezing | 172 (34.7) |
| Chest X-ray | |
| “Consolidation” | 391 (79) |
| “Other infiltrates” | 104 (21) |
| Para-pneumonic pleural effusion | 112 (22.6) |
| Uncomplicated para-pneumonic pleural effusion | 83 (16.8) |
| Complicated para-pneumonic pleural effusion | 29 (5.8) |
| Antibiotics during admission | 480 (97) |
| Admitted in PICU | 47 (9.5) |
| Continuous variables | Median (IQR) |
| Age (months) | 37 (18–66.3) |
| Days of axillar temperature ≥38°C at admission | 3 (2–5) |
| Transcutaneous oxygen saturation % (FiO2 0.21) | 94 (91–97) |
| Leucocytes/µl | 13,910 (9860–19,400) |
| Neutrophils/µl | 9260 (5800–14,200) |
| Lymphocytes/µl | 2450 (1500–4050) |
| Sodium (mmol/L) | 136 (135–138) |
| Albumin (g/dl) | 3.7 (3.3–4.1) |
| C reactive protein (mg/L) | 65 (26.9–155) |
| Procalcitonin (ng/ml) | 0.34 (0.09–1.8) |
| Test | N/global (%) |
|---|---|
| Blood culture | 489/495 (98.8) |
| PCR in blood for Streptococcus pneumoniae | 393/495 (79.4) |
| PCR in blood for other typical bacteria | 286/495 (57.8) |
| Serology for atypical bacteria at admission | 461/495 (93.1) |
| Serology for atypical bacteria in convalescence | 337/495 (68.1) |
| PCR for 16 viruses in nasopharyngeal aspirate | 459/495 (92.7) |
| PCR for atypical bacteria in nasopharyngeal aspirate | 441/495 (89.1) |
| Pleural fluid culture | 39/39 (100) |
| S. pneumoniae antigen in pleural fluid | 38/39 (97.4) |
At least one likely causative pathogen was identified in 262 patients (52.9%; Table 3A). Total viral detections in the NPA of 495 patients, including the likely and non-likely causative agents, are shown in Table S1. Considering only the likely causative pathogens, the most frequent etiological group was the viruses, which were detected in 155/262 (59.2%) children with CAP. Moreover, it was the single causative group of pathogens in 138/262 (52.7%) patients. RSV was the main viral agent and was detected in 60/262 (22.9%) patients. M. pneumoniae was the most frequent single causative agent that was detected in 78/262 (29.8%) patients. Of these 78 patients, M. pneumoniae was considered the main causative agent in only 76 (97.4%) patients, because it was present as a coinfection along with S. pneumoniae in the other two patients. Other AB, such as C. pneumoniae, were found in six additional patients (Table 3A). TyB was detected in 40/262 (15.3%) cases, with S. pneumoniae forming the majority (32/262, 12.2%). Coinfection with at least two different pathogens was observed in 22/262 (8.6%) patients having at least one identified pathogen. Most of the mixed infections included bacteria (17/22, 77%), mainly present along with viruses (14/22, 64%), and S. pneumoniae-M. pneumoniae coinfection (3/22, 13.7%). Only 5/22 (22.7%) of the cases showed virus-virus coinfection (Table 3B).
| A. Individual causative pathogens | |
|---|---|
| Pathogens | N (%) |
| Typical bacteria | 40 (15.3) |
|
32 (12.2) |
|
3 (1.1) |
|
3 (1.1) |
|
2 (0.8) |
| Atypical bacteria | 84 (32.1) |
|
78 (29.8) |
|
5 (1.9) |
|
1 (0.4) |
| Virus | 155 (59.2) |
|
60 (22.9) |
|
37 (14.1) |
|
42 (16%) |
|
16 (6.1) |
| B. Pathogens in co-infection | |
|---|---|
| Pathogens in co-infection | N (%) |
| Overall | 22 (8.6) |
| Typical bacteria-virus | 6 (2.4) |
|
2 (0.8) |
|
1 (0.4) |
|
1 (0.4) |
|
1 (0.4) |
|
1 (0.4) |
| M. pneumoniae-virus | 8 (3.1) |
|
4 (1.5) |
|
2 (0.8) |
|
1 (0.4) |
|
1 (0.4) |
| S. pneumoniae-M. pneumoniae | 3 (1.1) |
| Virus-virus | 5 (2.0) |
| PIV-HMPV | 2 (0.8) |
| PIV-RSV | 2 (0.8) |
| PIV-influenza | 1 (0.4) |
- Note: The individual pathogens amount to more than 100% because coinfections were found.
- Abbreviations: hMPV, human metapneumovirus; PIV, parainfluenza virus; RSV, respiratory syncytial virus.
In the CXR examinations, 391/495 (79%) patients showed consolidation and 104/495 (21%) had other infiltrates. In the group of patients with likely viral etiology, 89/138 (64.5%) had consolidation and 49/138 (35.5%) had other infiltrates. In the group with likely AB etiology, 72/82 (87.8%) had consolidation and 10/82 (12.1%) had other infiltrates, and 40/40 (100%) patients with likely TyB etiology had consolidation.
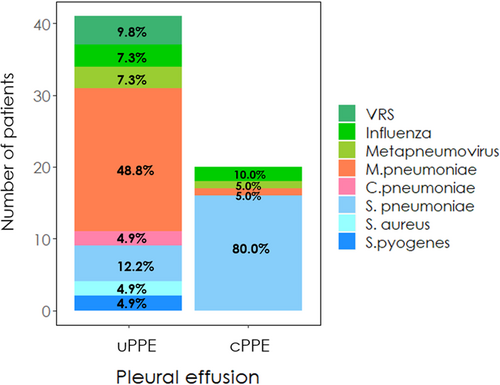
PPE was observed in 112/495 patients (22.6%), of which 29 (25.8%) had cPPE. Thoracentesis was performed in 39/112 (34.8%) patients. In patients with PPE, likely causative pathogens were detected in 61/112 (54.5%) patients: viruses in 12/61 (19.7%), AB in 23/61 (37.7%), and TyB in 26/61 (42.6%). Viruses and AB were mainly associated with ucPPE, and most cPPE (16/20, 80%) were caused by TyB (Figure S1). In total, 13/138 (16.6%) patients with viral etiology, 23/84 (27.3%) with AB etiology, and 25/40 (62.5%) with TyB etiology had PPE. The most common single microorganisms associated with PPE were S. pneumoniae (22/61, 36.1%; cPPE: 16/22, 72.7%) and M. pneumoniae (21/61, 34.4%; ucPPE: 20/21, 95.2%). All etiological agents associated with PPE are shown in Figure 1.
In total, 480/495 (97%) patients received antibiotics, mainly beta-lactams (440/495, 88.9%) and/or macrolides (113/495, 22.8%). Among the 495 patients, 269 (54.3%) required oxygen supplementation, 6 (1.2%) required mechanical ventilation, and 53 (10.7%) were admitted to the pediatric intensive care unit (PICU). There were no deaths reported. The median (IQR) days of admission were 5 (4–6) in children with viral etiology, 5 (4–7) in those with AB etiology, and 11.5 (6–16) in patients with TyB etiology.
Viral etiology was significantly more frequent in young patients (p < 0.001) and in those with low basal tcSatO2 (p = 0.001), high lymphocyte counts (p < 0.001), wheezing (p < 0.001), and other infiltrates on CXR examination (p < 0.001; Table 4 and Figure S2).
| Virus, N = 138 | Atypical bacteria, N = 84 | Typical bacteria, N = 40 | p | |
|---|---|---|---|---|
| Continuous variables | Median (IQR) | Median (IQR) | Median (IQR) | |
| Age (months) | 12.0 (3.0–33.0) | 60.0 (36.0–96.0) | 24.0 (12.0–60.0) | <0.001 |
| Days of fever at admission | 3.00 (2.00–5.00) | 5.00 (2.00–7.00) | 4.00 (2.00–5.00) | 0.002 |
| Maximum temperature (°C) | 39 (38.4–39.7) | 38.7 (38.0–39.5) | 39.3 (38.7–39.7) | 0.05 |
| Basal tcSatO2 at admission | 93.0 (90.0–95.0) | 95.0 (92.0–97.0) | 95.0 (93.0–97.0) | 0.001 |
| Leucocytes/µl | 11,030 (8292–15,100) | 11,300 (7802–16,782) | 17,935 (13,150–23,962) | <0.001 |
| Neutrophils/µl | 6700 (4100–9675) | 7435 (4608–11,950) | 13,150 (9900–19,522) | <0.001 |
| Lymphocytes/µl | 2705 (1900–4400) | 2345 (1400–3000) | 1884 (935–2875) | <0.001 |
| Hemoglobin (g/dl) | 11.6 (10.7–12.5) | 12.4 (11.6–13.3) | 11.4 (9.62–12.3) | <0.001 |
| C-reactive protein (mg/L) | 48.0 (22.2–116) | 44.5 (19.6–98.2) | 291 (236–349) | <0.001 |
| Procalcitonin (ng/ml) | 0.33 (0.11–1.19) | 0.11 (0.04–0.62) | 13.9 (1.75–41.5) | <0.001 |
| Albumin (g/dl) | 3.70 (3.40–4.10) | 3.60 (3.45–4.20) | 3.20 (2.50–3.50) | <0.001 |
| Sodium (mMol/L) | 137 (135–139) | 137 (135–138) | 135 (133–137) | 0.001 |
| Categorical variables | N (%) | N (%) | N (%) | |
| Male gender | 80 (58) | 41 (48.8) | 25 (62.5) | 0.325 |
| History of asthma or airway reactive disease | 19 (13.8) | 13 (15.5) | 5 (12.5) | 0.125 |
| Antibiotic before admission | 22 (15.9) | 28 (33.3) | 3 (7.5) | 0.001 |
| Wheezing | 74 (53.6) | 21 (25.6) | 5 (12.5) | <0.001 |
| Consolidation | 89 (64.5) | 74 (88.1) | 40 (100) | <0.001 |
| “Other infiltrates” | 49 (35.5) | 10 (11.9) | 0 (0.00) | |
| Pulmonary complications (PPE, abscess, necrotizing pneumonia, pneumothorax) | 13 (8.7) | 24 (27.9) | 26 (71.1) | <0.001 |
| PPE | 13 (9.4) | 23 (27.4) | 25 (65.7) | <0.001 |
| Uncomplicated PPE | 10 (7.2) | 22 (26.2) | 9 (22.5) | <0.001 |
| Complicated PPE | 3 (1.5) | 1 (1.2) | 16 (42.5) | |
| PICU admission | 12 (8.7) | 17 (20.2) | 15 (37.5%) | <0.001 |
- Note: For the purposes of this analysis, in coinfections of typical bacteria with other groups, typical bacteria were considered the main agent. In coinfections atypical bacteria-virus, atypical bacteria were considered the main agent.
- Abbreviations: CXR, chest X-ray; PICU, pediatric intensive care unit; PPE, para-pneumonic pleural effusion; tcSatO2, transcutaneous saturation of oxygen.
Multinomial univariable logistic regression revealed that patients with CAP caused by AB were more likely to have more days of fever before admission (odds ratio [OR]: 1.22 [1.09–1.36], p = 0.0004) and a lower maximum temperature (OR: 0.74 [0.57–0.96], p = 0.026) than those with CAP caused by viruses. As compared to patients with viral CAP, those with AB CAP had an almost three-time higher risk of presenting with chest pain (OR: 2.71 [1.06–6.92], p = 0.040), 85% lesser risk of presenting dyspnea (OR: 0.15 [0.08–0.28], p = 1.87 × 10−9), and were more likely to have received antibiotics before admission (OR: 2.60 [1.40–5.01], p = 0.003). Patients with AB CAP were more likely to have higher hemoglobin levels (OR: 1.61 [1.29–2.0], p = 2.4 × 10−5) than those with viral CAP (Figure 2).
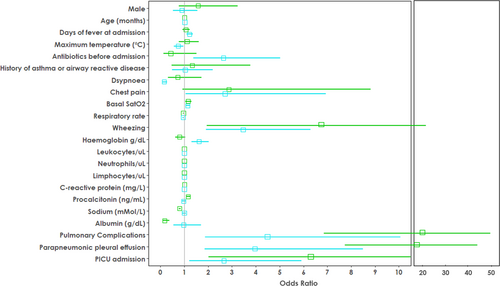
As compared to the patients with viral CAP, those with AB or TyB CAP were more likely to have higher basal tcSatO2 (ORatypical: 1.14 [1.05–1.23], p = .0.002; ORtypical: 1.17 [1.04–1.3], p = 0.007), lesser risk of presenting wheezing (ORatypical: 0.29 [0.16–0.52], p = 4.34 × 10−5; ORtypical: 0.12 [0.04–0.34], p = 3.8 × 10−5), higher leukocyte count (ORatypical: 1.00003 [1.00001–1.0001], p = 1.7 × 10−13; ORtypical: 1.0001 [1.0001–1.0001], p = 1.7 × 10−18), higher neutrophil count (ORatypical: 1.0001 [1.0001–1.0001], p = 1.2 × 10−7; ORtypical: 1.0001 [1.0001–1.0001], p = 1.7 × 10−18), and lower lymphocyte count (ORatypical: 0.99 [0.99–0.99], p = 4.56 × 10−6; ORtypical: 0.99 [0.99–0.99], p = 1.75 × 10−4; Figure 2).
Similarly, as compared to patients with viral CAP, those with bacterial CAP (AB or TyB) presented more frequent pulmonary complications (ORatypical: 4.97 [2.35–10.5], p = 2.7 × 10−5; ORtypical: 21.8 [8.97–52.99], p = 1.02 × 10−11) and PPE (ORatypical: 3.96 [1.84–8.49), p = 3.99 × 10−4; ORtypical: 19.5 [8.09–46.97], p = 3.52 × 10−11) and were more likely to be admitted to the PICU (ORatypical: 2.66 (1.2–5.91], p = 0.020; ORtypical: 6.3 [2.63–15.10], p = 3.52 × 10−5; Figure 2).
4 DISCUSSION
This study shows that viruses are the main microbiological agents associated with CAP in hospitalized pediatric patients in Spain. RSV was the most frequently detected microorganism, followed by AB, and M. pneumoniae was the most frequent etiological agent detected as a single causative agent.
The use of sensitive PCR assays for testing NPA specimens revealed a high prevalence of CAP caused by respiratory viruses and M. pneumoniae. Although the accuracy of upper respiratory tract samples to infer causality has been questioned, in the case of M. pneumoniae, several reports show a high correlation between detection by means of PCR and serology.4, 9, 18-20 For viruses, there are no reports showing such a correlation, and other ways to infer causality are currently unavailable for most clinical practitioners.
Viruses are considered the main causative agents of pediatric CAP worldwide. The reported rate of viral etiology is 34%–82%, and the results of our cohort were within this range, even though we used a very restricted definition of viral CAP.4, 8, 11, 18, 21-25 Only the following viruses mostly recognized as etiological agents of CAP were considered as the likely causative pathogens: RSV, hMPV, IV, and PIV. RSV is the most commonly detected viral agent in etiological studies of pediatric CAP, as also observed in our study.4, 8, 11, 18, 22-24 PIV, hMPV, and IV are also causative agents of CAP, but to a lesser extent.8, 13
The decision to exclude other viruses, such as hRV, ADV, EV, hCoV, and hBoV, from the viral CAP definition is controversial. At the time of the design, there were significant number of studies suggesting that controls have the same proportion of hRV, ADV, EV, hCoV, and hBoV as that of children with pneumonia.11, 12, 14 We attempted to differentiate between colonization and infection using quantitative molecular techniques and to establish cutoff points. However, this aim was beyond the scope of this study and could not be performed. Since it is impossible to distinguish whether the patients with the aforementioned viruses were carriers alone or if the pneumonia was actually caused by these viruses, we decided to exclude them.
The detection rate of all the viruses detected in the NPA samples was similar to that described previously.4, 11, 18, 21, 26 The causal role of hRV is controversial. Although the authors of the PERCH study suggested a probable causality, they recognized the limitations of the studies conducted to date to confirm this causal role.4 In our study, hRV was present in 25% of patients with CAP; therefore, if the causality were attributed to hRV, this agent would be considered the most common.
Our results revealed a rate of 8.6% of mixed infections, which is lower than that reported in recent years. Our low coinfection rate can be attributed to the low proportion of virus-virus coinfections (2%) as compared to that of mixed viral infections previously reported (20%–35%).8, 11, 21, 26, 27 This can be explained by the restrictive selection of pathogenic agents in our study. Most of the mixed infections in our study included bacteria-virus infections and dual bacterial infections (S. pneumoniae with M. pneumoniae). In the PERCH study, when only etiological agents of PF or lung aspirate were reported, there were only few mixed infection cases, mainly with bacteria-bacteria complexes and limited involvement of viruses.4 We believe that these data may question the very high rate of mixed infections commonly noted in pediatric CAP cases.
In our cohort, M. pneumoniae was the most frequent causal agent of pediatric CAP, accounting for nearly 30% of the cases. In another series, the prevalence of M. pneumoniae ranged from 1% to 38%.4, 7, 22, 26, 28-30 The differences in some large series may depend on the geographic variations.4-26 M. pneumoniae was associated with more days of fever and prescription of ambulatory beta-lactams. Therefore, pediatricians should consider M. pneumoniae as a likely etiological agent in children with CAP not responsive to beta-lactams, rather than considering it a failure of oral treatment. This is relevant because M. pneumoniae infection is often self-limiting.31 A more rapid diagnostic system for differentiating M. pneumoniae infections from TyB infections is required.
In this cohort, S. pneumoniae was found only in 15% of patients, as in other previous reports (5%–17.5%).23, 26 The diagnosis of pneumococcal CAP usually relies on culture tests or the presence of DNA in the blood and PF. We included the detection of S. pneumoniae antigen in PF in the workup.32 Better diagnostic performance can be accomplished using bronchoalveolar lavage or lung aspiration samples, but these procedures are considered too invasive in children not having a severe infection. The GABRIEL and PERCH studies suggested that some patients with a high density of S. pneumoniae in the nasopharynx might be infected by this microorganism, and they reported a frequency of pneumococcal etiology of 27% and 42%, respectively.4, 18 Some authors suggested testing the humoral response to S. pneumoniae. Subsequently, antibody response to S. pneumoniae was detected in 12%–53% of children25, 28, 30, 33; however, the specificity is low because of the potential detection of antibodies against colonizing pneumococci. Neither serology nor quantitative methods detecting high density of S. pneumoniae is widely available in clinical practice. The accurate diagnosis of pneumonia associated with S. pneumoniae and other TyB without invasive diagnostic methods is a challenging issue that requires better solutions.
PCR detection of S. pneumoniae and other bacteria in the blood deserves special attention. This detection has been usually considered demonstrative of the etiology. However, the PERCH findings are in contrast with this, since a similar proportion of S. pneumoniae using PCR was found in both cases and controls.4 We preferred to adhere to the usual interpretation of PCR of a TyB in the context of pneumonia as the most likely pathogen.32 The same occurred in two patients with non-typeable H. influenzae detection in blood with PCR. We included them in the TyB group, but this finding should be interpreted cautiously.
TyB are usually associated with features such as consolidation, PPE, and other pulmonary complications. Our study partially challenges some of these concepts. We found a significant proportion of patients with PPE in whom viruses and AB, particularly M. pneumoniae, were documented. Most of these were ucPPE. However, cPPE was mostly associated with TyB. We cannot attribute causality to microorganisms detected outside blood or PF with certainty, nor exclude TyB etiology if we do not detect them in these biological fluids, because of the low sensitivity of conventional cultures. Nonetheless, the fact that PPE associated with viruses and AB are usually ucPPE and the almost total absence of cPPE associated with these etiological groups indicates that even viruses and AB, and not just TyB, could be associated with ucPPE as has been suggested in others studies.34-37 Currently, based on our data, the most frequent cause of PPE is M. pneumoniae.
Another common concept challenged by this study is the association of consolidation with TyB. In our study, consolidation in the CXR was observed in all etiological groups, and not only with TyB. In contrast, other infiltrates were almost exclusively associated with viruses. Other features associated with viruses were low tcSatO2 and wheezing, similar to the findings of other studies.20, 38
The high proportion of viral etiology cases supports the idea that pediatric CAP treatment does not necessarily require antibiotics. Up to 97% of our patients received antibiotics, similar to that observed in other studies.25, 26, 33
In 2011, the British Thoracic Society guidelines for the management of CAP in children recommended the use of beta-lactam antibiotics in all children diagnosed with pneumonia, as bacterial pneumonia and viral pneumonia cannot be distinguished from each other reliably.39 A recent trial showed that the outcome on day 14 was similar for patients with WHO-defined pneumonia receiving either amoxicillin or placebo.40 Universal treatment with beta-lactams might not be justified by the current proportions of viral and AB pneumonia cases. We should consider clinical, radiographic, and biomarker data to ensure the judicious use of antibiotic therapy in pediatric CAP.
Our study has several limitations. TyB etiology may have been underdiagnosed because of the low sensitivity of conventional cultures and molecular tests in blood, as seen in other studies. Because of the restrictive definition of the likely viral etiology, it may have been underrepresented. Molecular techniques in the blood were only performed for TyB cases. The population of this study only included hospitalized patients; hence, the results cannot be representative of ambulatory CAP cases. The current coronavirus disease (COVID-19) pandemic caused by severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2) has affected millions of people worldwide, including children and adolescents.41 Pneumonia is one of the main manifestations of COVID-19, but it has distinctive clinical and inflammatory data. Our study was conducted before the pandemic. In the near future, pneumonia caused by SARS-CoV-2 will need to be excluded from the microbiological diagnosis. Our data will maintain the same validity in the remaining patients with CAP.
To summarize, CAP was mainly associated with viruses and AB in our study of hospitalized Spanish children and adolescents. M. pneumoniae was the most frequent causative agent. Viruses and AB can also cause PPE, but cPPE is mainly caused by TyB. Children and adolescents hospitalized with CAP do not always require antibiotics. The need for antibiotic treatment in children admitted with CAP must be individualized. Wheezing, presence of other infiltrates on CXR, and other signs could indicate viral etiology of CAP and help the clinician improve the antibiotic stewardship.
ACKNOWLEDGMENTS
The authors thank all the patients and their families for their participation in this study and the members of the medical and nursing staff who took care of them. Places of primary research: Pediatrics Department, Hospital Universitario Ramón y Cajal, Universidad de Alcalá, Madrid, Spain; Pediatrics Department, Hospital Universitario Infanta Sofía, San Sebastián de los Reyes, Madrid, Spain; Pediatrics Research Group. Universidad Europea de Madrid, Madrid, Spain. This study was supported by: Project PI17/01458, from the Instituto de Salud Carlos III (Ministry of Economy, Industry, and Competitiveness) and co-funded by the European Regional Development Funds. Instituto Ramón y Cajal de Investigación Sanitaria (IRYCIS) for research work by emerging research groups as part of the project “Etiology of Community-Acquired Pneumonia in Hospitalized Children” (PCAPE), in collaboration with the Microbiology Department of Hospital Universitario Ramón y Cajal. Reference PCAPE 2011_0025. Register 320/11. Research Project of Universidad Europea de Madrid (2017/UEM03). Research Grant of the Instituto de Investigación Hospital 12 de Octubre (i+12) number: AY191212-1. The funders had no role in study design, data collection, and analysis, the decision to publish or the preparation of the manuscript.
PCAPE AND VALS DANCE WORKING GROUPS
Raquel Buenache (Hospital Universitario Ramón y Cajal), Nathalia Gerig (Hospital Universitario Ramón y Cajal), Adelaida Lamas (Hospital Universitario Ramón y Cajal), Sinziana Stanescu (Hospital Universitario Ramón y Cajal), Pablo Morillo (Hospital Universitario La Paz), Rut del Valle (Hospital Universitario Infanta Sofía), Julia Yebra (Hospital Universitario Infanta Sofía), Rosa Batista (Hospital Universitario Infanta Sofía), Teresa Raga (Hospital Universitario Infanta Sofía), María García-Baró (Hospital Universitario Infanta Sofía), Magdalena Hawkins (Hospital Universitario Infanta Sofía, Universidad Europea), Alfonso Cañete (Hospital Universitario Infanta Sofía), Luis Prieto (Hospital Universitario 12 de Octubre), Lidia Oviedo (Hospital Universitario 12 de Octubre), Daniel Blázquez (Hospital Universitario 12 de Octubre), Manuel Gijón (Hospital Universitario 12 de Octubre), Lucía Figueroa (Fundación para la Investigación Biomédica del Hospital 12 de Octubre), Raquel Ramos Corral (BR Salud, Hospital Universitario Infanta Sofía), Nazaret del Amo (BR Salud, Hospital Universitario Infanta Sofía), María José Cilleruelo (Hospital Universitario Puerta de Hierro), María Luz Golmayo (Hospital Universitario Puerta de Hierro), María Isabel Sánchez (Hospital Universitario Puerta de Hierro), Beatriz Soto (Hospital Universitario de Getafe), Sara Guillén (Hospital Universitario de Getafe), David Molina (Hospital Universitario de Getafe), Mercedes Alonso-Sanz (Hospital Universitario Niño Jesús), Elvira Martín (Fundación Jiménez Díaz, Madrid), Cristina Calvo (Hospital Universitario La Paz), Ana Méndez-Echeverría (Hospital Universitario La Paz), Teresa del Rosal (Hospital Universitario La Paz), María Pilar Romero (Hospital Universitario La Paz), Alfonso Rodríguez-Albarrán (Hospital del Sureste), Manuel Imaz (Hospital Universitario de Basurto, Bilbao), Esther Casado Verrier (Hospital de Villalba), José-Tomás Ramos (Hospital Clínico San Carlos. Madrid).
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Enrique Otheo, Mario Rodríguez, and Alfredo Tagarro conceptualized and designed the study. Enrique Otheo, Alfredo Tagarro, Cinta Moraleda, Sara Domínguez-Rodríguez, and Mar Santos performed data management. Sara Domínguez-Rodríguez and Mar Santos performed the statistical analysis. Enrique Otheo, Alfredo Tagarro, and Sara Domínguez-Rodríguez drafted the manuscript. Enrique Otheo, Alfredo Tagarro, Mario Rodríguez, and Juan Carlos Galán were involved in the preparation and review of the final manuscript. All other co-authors enrolled participants and participated in the collection of data. All co-authors participated and were involved in the critical review of the final manuscript.
Open Research
DATA AVAILABILITY STATEMENT
Data openly available in a public repository that issues data sets with DOIs.




