Expanding the OMOP Common Data Model to Support Perinatal Research in Network Studies
Funding: This project has received support from the European Health Data and Evidence Network (EHDEN) project. EHDEN received funding from the Innovative Medicines Initiative 2 Joint Undertaking (JU) under grant agreement No 806968. The JU receives support from the European Union's Horizon 2020 research and innovation programme and EFPIA. The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript. Norwegian registry data were harmonized into OMOP-CDM supported by a European Health Data & Evidence Network (EHDEN) project grant.
ABSTRACT
Objectives
The Observational Medical Outcomes Partnership common data model (OMOP-CDM) is a useful tool for large-scale network analysis but currently lacks a structured approach to pregnancy episodes. We aimed to develop and implement a perinatal expansion for the OMOP-CDM to facilitate perinatal network research.
Methods
We collaboratively developed a perinatal expansion with input from domain experts and stakeholders to reach consensus. The structure and vocabularies followed the OMOP-CDM ontological framework principles. We tested the expansion using SIDIAP and Norwegian databases. We developed a diagnostics package for quality control assessment and conducted a descriptive analysis on the captured perinatal data mapped to the OMOP-CDM.
Results
The perinatal expansion consists of a pregnancy table and an infant table, each with required and optional variables incorporated into standardized vocabularies. Quality assessment of the perinatal expansion table in SIDIAP and Norwegian databases demonstrated accurate capture of perinatal characteristics. Descriptive analysis measured the number of pregnancies (SIDIAP: 646 530; Norway: 746 671), pregnancy outcomes (e.g., 0.5% stillbirths in SIDIAP and 0.4% in Norway), gestational length (median [IQR] in days, SIDIAP: 273 [56–280]; Norway: 280 [273–286]), number of infants (Norway: 758 806), and birth weight (median [IQR] in grams, Norway: 3520 [3175–3860)], among other relevant variables.
Discussion and Conclusion
We developed and implemented a perinatal expansion that captures important variables for perinatal research and allows interoperability with existing tables in the OMOP-CDM, which is expected to facilitate future network studies. The publicly available diagnostics package enables testing the implementation of the extension table and the quality and completeness of available data on pregnancy and pregnancy-related outcomes in databases mapped to the OMOP CDM.
Summary
- We collaboratively developed a perinatal expansion for the OMOP-CDM and a diagnostics package for quality control assessment.
- The perinatal expansion consists of a pregnancy table and an infant table that capture relevant variables for observational perinatal research.
- The perinatal expansion tables allow interoperability with existing tables in the OMOP-CDM.
- The perinatal expansion was successfully implemented in two large European databases and is ready to be used by other databases mapped to the OMOP-CDM with pregnancy and mother-child related information.
- The perinatal expansion is expected to facilitate future perinatal network research using the OMOP-CDM.
1 Background and Significance
The perinatal period, consisting of pregnancy, delivery and postpartum, is an essential time for the health of the mother and child. Understanding various conditions and risk factors in this period is vital, also for long-term health outcomes [1]. Pregnant individuals are often excluded from clinical trials due to ethical reasons. In this scenario, observational health data offers a great opportunity. Many observational healthcare databases, such as electronic health records databases or national patient registries, contain detailed data related to the perinatal period and can be a valuable source of information for perinatal research [2], as demonstrated in recent studies filling urgent evidence gaps related to COVID-19 [3-6].
The Observational Health Data Science and Informatics (OHDSI) initiative [7] maintains the Observational Medical Outcomes Partnership common data model (OMOP-CDM), a widely used data model for harmonizing observational healthcare data in a standardized format. As of 2024, more than 544 databases worldwide were mapped to the OMOP-CDM [8]. The OMOP-CDM is the backbone of a suite of OHDSI tools that offers numerous advantages, including enhanced interoperability among healthcare systems and data sources, streamlined analytics, and reproducibility of research studies through the use of consistent data representation schemes and study protocols [9]. However, the current structure of the OMOP-CDM is limited to define pregnancy episodes including important features associated with the perinatal period, such as gestational length, the number of fetuses associated with a pregnancy. Thus, it is challenging to perform multi-database network studies on perinatal pharmacoepidemiology using databases mapped to the OMOP-CDM. A number of algorithms to identify pregnancy episodes from claims and electronic health records (EHR) data have been previously developed [10-12], but a standardized structure for its representation in the OMOP-CDM and OHDSI vocabulary that is applicable for claims, EHRs and registry data is not yet available.
Adding an expansion to the OMOP-CDM could facilitate researchers to analyze data related to pregnancies, deliveries, and neonatal outcomes by clearly identifying pregnancy. In addition, the expansion would allow for more accurate and efficient analysis across different healthcare systems and databases, thus, greatly enhancing network studies on maternal–infant health, which will ultimately lead to improved care and outcomes for pregnant people and their infants.
Here, we describe the development of a perinatal expansion to the OMOP-CDM and test its implementation in two use cases. This expansion aims to represent pregnancy episodes and its associated information in a structured way. The addition of a perinatal expansion to the OMOP-CDM aims to enhance and facilitate network perinatal studies within the observational health data partners mapped to the OMOP-CDM provided that detailed perinatal information can be derived from the data source.
2 Methods
2.1 Perinatal Expansion Conceptualization
The proposed perinatal expansion to the OMOP-CDM was collaboratively developed, with input from perinatal domain experts, epidemiologists, data analysts, and other stakeholders who provided feedback on the proposed expansion through several rounds of review and discussions to reach consensus. The aim of this collaborative process was to ensure that the proposed expansion reflected current clinical and research needs in the perinatal field, as well as complied with relevant considerations related to data standardization and interoperability.
The proposed expansion structure and concepts were developed under the established OMOP-CDM ontological framework principles of using unique standard concepts (1) that are precise, sufficiently granular, unambiguous, and non-redundant, (2) assigned to specific domains, (3) comprehensive in their coverage of the semantic space, (4) represented in an ontology allowing for hierarchy and relationships between concepts to be integrated into existing ontologies [13].
2.2 Perinatal Expansion Implementation
2.2.1 Data Sources
We piloted the perinatal expansion in two settings: SIDIAP database (The Information System for Research in Primary Care; Spain) and Norwegian linked registries data, managed by the University of Oslo (UiO). We chose these databases to test the applicability of the pregnancy expansion to different types of data sources. We selected SIDIAP electronic health records (EHR) database and the Norwegian national health registries as use cases to test the applicability of the pregnancy expansion due to their unique yet complementary characteristics in terms of data provenance, data types, and data granularity; their rich and reliable perinatal data; their demonstrated history of use in perinatal research [14, 15]; and their active participation in international research networks (e.g., European Health Data Evidence Network [EHDEN], Data Analysis and Real-World Interrogation Network [DARWIN EU], and OHDSI).
Spain has a universal public healthcare system that is decentralized into its 17 autonomous communities. SIDIAP is a population-wide EHR database from Catalonia, Spain [16]. It includes pseudo-anonymized routinely collected records from > 8 million people collected from 328 primary care centers since 2006 with updates every 6 months. The population included in the database represents 75% of the Catalan population and is representative of the overall Catalan population in terms of sex, age and geographic distributions. SIDIAP data has been mapped to the OMOP CDM [17] and includes high-quality data on disease diagnoses, drug prescription and dispensations, laboratory tests, clinical measurements, lifestyle and demographic characteristics, among others. In addition, it includes detailed information related to the perinatal period for approximately 40 000 deliveries per year.
Norway has a universal public healthcare system covering all inhabitants, approximately 5.4 million people. Linkage of registry data is done using the unique personal identification number assigned at birth or immigration for all residents in Norway [18]. The data that UiO mapped onto the OMOP CDM includes high-quality data on disease diagnoses from registries covering primary and secondary care, drug dispensations, vaccine administration, and positive tests for communicable diseases, covering the entire Norwegian population from 2008 to 2021. These registries include the Norwegian Prescription Database (NorPD) [19], the national vaccination registry (SYSVAC) [20], the Norwegian Surveillance System for Communicable Diseases (MSIS) [21], the Norwegian Patient Registry (NPR), and the Norwegian Control and Payment of Health Reimbursements Database (KUHR) [22]. Importantly, it includes detailed information related to all pregnancies lasting more than 12 weeks of gestation in Norway (information on the mother, father, and child), approximately 60 000 deliveries per year from the Medical Birth Registry of Norway (MBRN) [23].
2.2.2 Mapping Source Data to the OMOP CDM Perinatal Expansion
We followed the Extract, Transform and Load (ETL) process to map source data into the perinatal expansion [24]. This process refers to the steps necessary to convert source data from its original form and structure into a standardized format required within the OMOP-CDM. The ETL process involved data and CDM experts to design the ETL, field and medical experts to create the code mappings (i.e., mapping terminology in the source data into standard OMOP vocabularies), and technicians to implement the ETL in source data. In the ETL process, we first extracted the data from its original form in the source database. Second, we transformed the data creating the code mappings separately in SIDIAP and the Norwegian databases. Third, we loaded the transformed data into the OMOP-CDM perinatal expansion tables.
2.2.3 Quality Control Assessment
We developed a publicly available diagnostics R package (PET Diagnostics; available at https://github.com/oxford-pharmacoepi/PETDiagnostics) to ensure that the perinatal expansion for the OMOP CDM was implemented correctly in each database [25]. This diagnostics package includes a set of tests and queries designed to assess the quality and completeness of the perinatal data captured in each table. The results regarding quality and completeness will reflect potential issues in both the PET and the source data. Complete information on the diagnostics package is found in Methods S1 and Table S1.
2.2.4 Descriptive Analyses
We conducted a descriptive analysis on the information captured in the perinatal expansion in both data sets for the period 01/01/2006–31/12/2020 for SIDIAP and 01/01/2008–31/12/2020 for Norway.
3 Results
3.1 Perinatal Expansion Conceptualization
The expansion consists of a pregnancy table and an infant table. Each table includes a set of required and optional variables corresponding to each pregnancy episode. A pregnancy episode is considered as the period comprised between the pregnancy start (i.e., the date of last menstruation) and the pregnancy end (i.e., the date of delivery, stillbirth, miscarriage, or termination of pregnancy). Every pregnancy episode in the pregnancy table is identified by a unique ID that is linked to the pregnant person's ID. The pregnancy table is designed to identify pregnancy episodes with defined start dates, end dates and gestational length, and the most relevant information including pregnancy outcomes, mode of delivery, and whether it is a singleton or a multiple pregnancy. The expansion also comprises more detailed information on the current pregnancy episode (e.g., number of fetuses) and information related to previous pregnancies (e.g., parity). The infant table is designed to capture information on each newborn infant. Every infant in the infant table is identified by a unique ID that is linked to the corresponding pregnancy ID. The identification of each infant is especially relevant in the case of pregnancies with more than one infant, in which different birth outcomes can occur and infant characteristics are expected. The table includes relevant information including birth outcomes, birth weight, congenital malformations, and APGAR scores, among others. Full structure and content of the perinatal expansion is described in Table 1 for pregnancies and Table 2 for the infants. ETL specifications for the pregnancy and infant expansion tables are presented in Tables S2 and S3.
| CDM field | Description |
|---|---|
| Required fields | |
| person_id | Unique identifier of the pregnant person |
| pregnancy_id | Unique identifier of each pregnancy episode |
| pregnancy_start_date | Date when the pregnancy episode started (based on ultrasound estimations or calculated from last menstrual period if ultrasound information is missing) |
| pregnancy_end_date | Date when the pregnancy episode ended (based on the date of the pregnancy outcome) |
| gestational_length_in_day | Length of gestation in days |
| pregnancy_outcome | Outcome of the pregnancy: livebirth, miscarriage (< 20 weeks), stillbirth (≥ 20 weeks), elective termination of pregnancy, discordant (different outcomes in multiple pregnancies), unknown |
| pregnancy_mode_delivery | How the delivery was initiated: vaginal, c-section, unknown |
| pregnancy_single | Single pregnancy: yes if single, no if multiple, unknown |
| Optional fields | |
| pregnancy_folic | Reported using folic acid in recommended period: yes, no |
| pregnancy_number_fetuses | Number of fetuses in the given pregnancy |
| pregnancy_art | Received Assisted Reproductive Technology treatment for the given pregnancy: yes, no |
| pre_pregnancy_bmi | Pre-pregnancy BMI |
| pregnancy_marital_status | Marital status of the pregnant person at delivery: single, married/cohabiting, other |
| pregnancy_number_liveborn | Number of liveborns in the given pregnancy |
| prev_pregnancy_parity | Previous pregnancies with ≥ 20 weeks of gestation: nulliparous, multiparous |
| prev_pregnancy_gravidity | Number of previous pregnancies regardless of gestational length |
| prev_livebirth_number | Number of previous livebirths |
| prev_still_misc_number | Number of previous stillbirths and miscarriages |
| prev_top_number | Number of previous terminations of pregnancy, regardless of gestational length |
| pregnancy_outcome_source_value | Value of the pregnancy outcome in the source data |
| pregnancy_mode_delivery_source_value | Value of the pregnancy mode of delivery in the source data |
| CDM field | Description |
|---|---|
| Required fields | |
| pregnancy_id | Unique identifier of each pregnancy episode |
| infant_id | Unique identifier of each infant in the given pregnancy |
| Optional fields | |
| birth_outcome | Birth outcome: livebirth, miscarriage (< 20 weeks), stillbirth (≥ 20 weeks), elective termination of pregnancy |
| birth_weight | Birth weight measured in grams |
| birth_con_malformation | Born with a congenital malformation: yes, no |
| birth_sga | Small for gestational age of the newborn: yes, no |
| birth_fgr | Fetal growth restriction of the newborn: yes, no |
| birth_apgar | Score obtained from the Apgar test 5 min after birth |
3.2 Perinatal Expansion Implementation
The pregnancy table was successfully implemented in SIDIAP and Norwegian databases, while the infant table was only implemented in Norwegian data. The SIDIAP source data structure does not currently include mother–child linkage records, but these are in the process of being added. When they are available, the infant table will be implemented for SIDIAP as well. We were able to map all required variables from source data. Complete information of code mappings for the required variables in SIDIAP and Norwegian databases are presented in Table S4. For example, pregnancy start date was mapped from the recorded date of last menstruation corrected from ultrasound in SIDIAP and in Norway it was newly created from subtracting the recorded gestational length (ascertained mainly by ultrasound) to the recorded pregnancy end date. Pregnancy end date was mapped from the date of delivery or from the date of closing of the pregnancy record when the date of delivery was not available in SIDIAP and from the recorded pregnancy end date in Norway (Table S4). The quality assessment of the developed expansion tables was successful for both databases (Table S5). The expansion captured perinatal characteristics with no missing data in all required variables from both tables. The diagnostics package identified 22.9% and 0.4% unknown values for gestational length in SIDIAP and Norway, respectively (Table S5), which were the same as in the source data. There were 0.9% of records in SIDIAP and 0.4% in Norway that had a pregnancy end date on or before the start date. Ninety-eight percent of pregnancies in SIDIAP and 100% of pregnancies in Norway had plausible matches of pregnancy outcomes (e.g., livebirth) with mode of delivery (e.g., vaginal or C-section records) (Table S5).
3.3 Population Characteristics
A total of 646 530 pregnancies from 426 318 pregnant individuals were identified in SIDIAP and 746 671 pregnancies from 465 029 pregnant individuals were identified in Norway using the pregnancy table in the OMOP converted data (Table 3). Of all pregnancies, 0.5% in SIDIAP and 0.4% in Norway ended in stillbirths (≥ 20 weeks of gestation). C-section was performed in 32.9% (SIDIAP) and 7.1% (Norway) of pregnancies. Pregnant individuals had a mean pre-pregnancy BMI of 24.8 (SD = 4.9) in SIDIAP and 24.5 (SD = 4.9) in Norway. Additionally, infants from all pregnancies recorded in the pregnancy table (n = 746 671) were identified in the separate infant table in Norway. Of those infants, 98.9% were born alive and with a median birth weight of 3520 g (IQR = 685) and median APGAR score of 10 at 5 min after delivery (IQR = 1) (Table 4).
| SIDIAP | Norwegian databases | |||
|---|---|---|---|---|
| Pregnancy characteristics | Missing/unknown | Pregnancy characteristics | Missing/unknown | |
| Number of pregnancies, N | 646 530 | 0 (0%) | 746 671 | 0 (0%) |
| Pregnant individuals, N | 426 318 | 0 (0%) | 465 029 | 0 (0%) |
| Pregnancy identification period (from start dates) | 02/01/2006–31/12/2020 | — | 01/01/2008–31/12/2020 | — |
| Gestational length of infant, days, median (IQR) | 273 (56–280) | 0 (0%) | 280 (273–286) | 0 (0%) |
| Pregnancy outcome, n (%) | 10 309 (1.6%) | 224 (0%) | ||
| Livebirth | 581 127 (91.3) | 738 401 (98.9) | ||
| Miscarriagea (< 20 w) | 46 996 (7.4) | 1343 (0.2) | ||
| Stillbirth (≥ 20 w) | 3069 (0.5) | 3024 (0.4) | ||
| Elective termination of pregnancya | 5029 (0.8) | 3664 (0.5) | ||
| Discordant outcome (in multiple pregnancies) | — | 15 (0.0) | ||
| Mode of delivery, n (%) | 0 (0%) | 0 (0%) | ||
| Vaginal | 434 135 (67.1) | 693 852 (92.9) | ||
| C-section | 212 395 (32.9) | 52 819 (7.1) | ||
| Single pregnancy, n (%) | 645 336 (99.8) | 0 (0%) | 734 413 (98.4) | 0 (0%) |
| Marital status, n (%) | 646 530 (100%) | 40 (0%) | ||
| Single | — | 39 747 (5.3) | ||
| Married | — | 328 435 (43.9) | ||
| Cohabiting | — | 367 174 (49.2) | ||
| Other/mixed | — | 2851 (0.4) | ||
| Number of fetuses, median (min, max) | 1 (1, 9) | 0 (0%) | 1 (1, 4) | 0 (0%) |
| Number of liveborns, median (min, max) | — | 646 530 (100%) | 1 (1, 4) | 0 (0%) |
| Parity, n (%) | 0 (0%) | 0 (0%) | ||
| Nulliparous | 258 712 (40) | 429 973 (57.6) | ||
| Multiparous | 387 818 (60) | 316 698 (42.4) | ||
| Gravidity, median (min, max) | 1 (0, 27) | 0 (0%) | 1 (0, 4) | 0 (0%) |
| Pre-pregnancy BMI, mean (SD) | 24.8 (4.9) | 61 846 (9.6%) | 24.5 (4.9) | 227 129 (32%) |
- a All pregnancies ending after gestational week 12 (GW12) are notifiable to the Norwegian birth registry, including terminations after week 12 requiring approval. Miscarriages prior to GW12 are not reported in the Norwegian birth registry.
| Infant characteristics | Missing/unknown | |
|---|---|---|
| Pregnancies, N | 746 671 | 0 (0%) |
| Infants, N | 758 806 | 0 (0%) |
| Birth outcome, N (%) | 251 (0%) | |
| Livebirth | 750 365 (98.9) | |
| Miscarriagea (< 20 w) | 1389 (0.2) | |
| Stillbirth (≥ 20 w) | 1447 (0.4) | |
| Elective termination of pregnancya | 3112 (0.5) | |
| Birth weight, gram, median (IQR) | 3520 (31753860) | 4359 (0.6%) |
| Born with congenital malformation, yes N (%) | 34 093 (4.5) | 0 (0) |
| APGAR score (5 min), median (IQR) | 10 (9–10) | 4796 (0.6%) |
- a All pregnancies ending after gestational week 12 (GW12) are notifiable to the Norwegian birth registry, including terminations after week 12 requiring approval. Miscarriages prior to GW12 are not reported in the Norwegian birth registry.
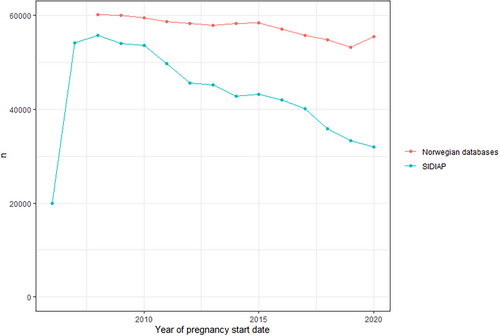
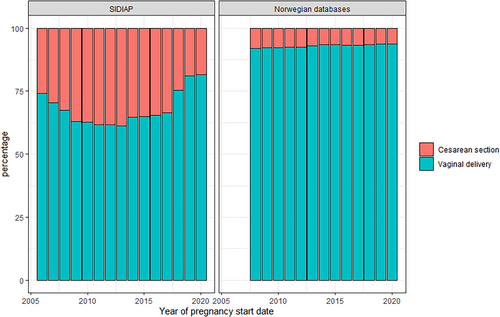
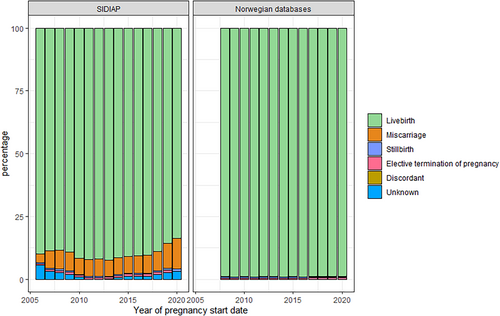
The number of new pregnancies per calendar year decreased during the study period. This is consistent with national birth statistics for Spain [26] and Norway [27, 28] (Figure 1). The number of pregnancies in SIDIAP dropped from 54 091 in 2007 to 31 994 in 2020, and in Norway from 60 041 in 2008 to 55 407 in 2020. In SIDIAP, the number of pregnancies captured in the first year of data availability of the database (2006) was lower than expected from national statistics, but was consistent to available source data [26]. While in Norway the proportion of C-section deliveries slightly decreased during the study period (7.9% in 2008 to 6.3% in 2020), the number of C-section performed in SIDIAP increased to 38.8% of all pregnancies in 2013 then began to decrease to 18.4% of pregnancies in 2020 (Figure 2). Pregnancy outcomes in Norway remained stable throughout the study period, with low numbers of miscarriages, elective terminations of pregnancy and stillbirths after gestational week 12. In SIDIAP, the number of miscarriages increased up to 12% in 2020 (Figure 3). Of note, the number of miscarriages in SIDIAP is expected to be higher compared to Norway, because in Norway, only miscarriages occurring after week 12 of gestation are recorded. Figure 4 represents the distribution of gestational length (in weeks) for each pregnancy outcome, showing that most terminations of pregnancy occurred during the first and second trimesters. As expected, and as a validation of both gestational length and pregnancy outcome variables, there were no miscarriages recorded with gestational length greater than 20 weeks and no stillbirths less than 20 weeks. In SIDIAP, a low number of pregnancies had unknown values for pregnancy outcome but did include a record of gestational length. Of those, a notable proportion had gestational length around 40 weeks of gestation, which is the standard length of term pregnancies.
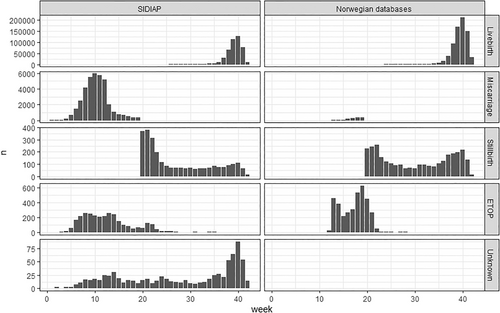
Distribution of gestational length in weeks per pregnancy outcome.
ETOP: Elective termination of pregnancy. Unknown: Unknown values that are mapped to 0. All pregnancies ending after gestational week 12 (GW12) are notifiable to the Norwegian birth registry, including terminations after week 12 requiring approval. Miscarriages prior to GW12 are not reported in the Norwegian birth registry.
4 Discussion
We have developed a perinatal expansion for the OMOP-DCM using standardized vocabularies and implemented it in two large databases in Europe. The implementation in these use cases allowed us to identify and characterize approximately 1.4 million pregnancies from 2006 to 2020 from both databases. The expansion captures important variables for observational perinatal research including pregnancy start and end dates, pregnancy outcomes, mode of delivery, and whether a pregnancy was singleton or multiple. It also captures valuable information related to each infant. By linking each pregnancy and infant to a person ID, the expansion tables allow interoperability with existing tables in the OMOP-CDM. Also, it enables a direct mother–infant linkage. The adoption of this expansion within other databases in the OMOP-DCM will facilitate collaborative network studies on perinatal research among diverse data partners.
EHDEN [29] and OHDSI [7] encompass a large network of observational health databases mapped to the OMOP-CDM, including electronic health records, registries, or claims databases. Many of those, including SIDIAP and Norwegian databases, contain relevant perinatal-related data that have not previously been used for federated network analysis and could benefit from including the perinatal expansion to their data. To date, researchers within the OHDSI and EHDEN networks have been limited in their ability to conduct multi-database network studies with perinatal data given the complexity of identifying pregnancy episodes and relating them to relevant data in other tables in the OMOP-CDM [10, 30]. Previous studies have developed algorithms to infer pregnancy episodes and pregnancy outcomes from source data, since direct measures of gestational length or of some pregnancy outcomes (e.g., stillbirths) are typically not available in a number of observational databases [10, 12, 30-37]. These algorithms are mostly needed to infer pregnancy start and end dates among databases that do not contain such dates nor gestational length [11]. Our perinatal expansion can be directly implemented among those databases with available pregnancy episodes identified in their source data and can complement those databases where an application of a pregnancy algorithm is needed as a first step. The perinatal expansion provides the harmonized structure to enable pregnancy episodes and related outcomes to be identified and analyzed at a granularity that is suitable for identifying reliable pregnant cohorts and facilitates conducting observational perinatal research.
A key part of validating and ensuring the robustness of this expansion involved applying it to real-world data sources. By transforming the source data from each data source into the CDM, we could verify that the proposed pregnancy and infant tables in the CDM expansion could capture relevant data elements (e.g., gestational age, maternal characteristics, birth outcomes) across different healthcare systems and data provenance. The implementation and testing of the perinatal expansion in SIDIAP and Norway databases demonstrated a successful representation of perinatal data among EHR and national registry data, showing its flexibility while maintaining data comparability. In the OMOP-CDM, the standardized vocabulary was limited for some pregnancy characteristics. Existing standardized concepts in medical vocabularies, such as ICD or SNOMED, are limited in their level of granularity (e.g., smoking status before and during pregnancy, previous miscarriages, or stillbirths). This required creating custom concepts for the perinatal expansion that were not supported but were considered necessary for a concise representation of the pregnancy characteristics.
Our quality control assessment demonstrated that it is possible to implement the perinatal expansion across diverse databases to develop perinatal network studies in observational health databases mapped to the OMOP-CDM. Required variables were 100% captured in both SIDIAP and Norwegian databases, and the expansion captured a substantial percentage of complementary variables, such as those related to previous pregnancies, that can enrich perinatal studies. Via the pregnant person's ID, the expansion allows linkage between the perinatal expansion tables and existing OMOP-CDM tables (i.e., person, observation, condition occurrence, drug exposure, etc.). This facilitates a wide range of much needed future network studies integrating perinatal data. The incorporation of the expansion in these two databases that actively participate in international research networks serves as an example on its potential and promotes the scalability of implementing the expansion into other data sources. Pregnant people are often excluded from trials for ethical and safety concerns, and as result, there are limited comprehensive studies on vaccine coverage, drug or vaccine safety and effectiveness, and risk of comorbidities during or before pregnancy on perinatal outcomes or on long-term offspring health. This represents a significant clinical gap that needs to be addressed. Current growing initiatives such as the DARWIN EU [38] can greatly benefit from this expansion as they leverage the OMOP-CDM to generate timely and reliable evidence and could provide valuable evidence on the use, safety, and effectiveness of medicines during pregnancy.
OHDSI comprises a network of collaborators and experts in the field of observational healthcare research and a dedicated working group on pregnancy and reproductive health research that can provide valuable contributions to further expand our work. The collective expertise within the OHDSI community can ensure that the perinatal expansion remains current, aligning with emerging research needs and evolving data standards. Especially, future work could focus on the use of the recently added episode and episode_event tables in the pregnancy context.
The present study contributes to the field in several ways. By providing a clear structure for standardized data integration, it facilitates network studies using perinatal data. This can promote collaboration across data partners leading to more comprehensive and robust studies, such as previously successful collaborative projects (e.g., CHARYBDIS [39]), that provided insightful and timely evidence to inform the COVID-19 response. Studies of a similar magnitude within the perinatal field using the OMOP-CDM have not been yet done and could greatly benefit the field. The successful implementation of the expansion in two large databases and the allowance to interact with existing tables within the CDM demonstrates the feasibility and practicality of incorporating such structure and vocabularies into an existing CDM. The compatibility with the established framework suggests that the expansion can be readily adopted by other researchers and institutions using the OMOP-CDM. Ultimately, it has practical implications for healthcare institutions and policymakers interested in leveraging the OMOP-CDM for perinatal research.
We acknowledge this study has limitations. First, we proposed the current tables to be an OMOP-CDM expansion. However, expansion tables are not incorporated into the common data model and thus they are not yet supported by some OHDSI tools such as ATLAS. Second, the expansion requires a set of essential variables to be included for its implementation, thus limiting the adoption in databases without data on all required variables. Third, the expansion may not be seamless in other settings. Different sources such as claims databases may have unique structures or technical limitations that could require additional adaptations of their source data, such as implementation of a claims-specific pregnancy episode detection algorithm [10, 40] and mother–infant linkage algorithm [40], prior to implementation.
5 Conclusion
We developed a perinatal expansion that will standardize perinatal data with a high level of granularity in the OMOP-CDM, thereby facilitating future network perinatal studies. The perinatal expansion tables were successfully implemented in two large European databases and are ready to be used by other databases mapped to the OMOP-CDM with pregnancy and mother–child related information. This expansion can serve as a valuable resource for researchers working in perinatal epidemiology using OMOP mapped data. Our publicly available PETDiagnostics R package enables testing the implementation of the pregnancy expansion as well as the quality and completeness of pregnancy-related data in databases mapped to the OMOP-CDM.
Author Contributions
A.A., N.T.H.T., E.B., T.B., E.H., D.R.M., H.N., T.D.-S. conceived and designed the expansion. N.T.H.T., S.F.-B., C.R., E.S. implemented the expansion into the databases. T.B., E.B. developed the diagnostics package and analytic codes. A.A., N.T.H.T., T.B. analyzed the results. A.A. wrote the first draft of the manuscript. All authors reviewed, interpreted the results, contributed to revisions, and read and approved the final version.
Acknowledgments
We thank Fabian Leonardo Martinez Bernal and Luigi Maglanoc at the IT Department at the University of Oslo and Jared Houghtaling from edenceHealth for support related to IT-infrastructure hosting Norwegian data.
Ethics Statement
This study was approved by the Clinical Research Ethics Committee of the IDIAPJGol (project code: 4R21/042), by the Regional Committee for Research Ethics in Norway (approval numbers: 155294/REK Nord and 2018/140/REK Sør Øst) and the Data Protection Officer at the University of Oslo (approval numbers: 523275 and 58 033).
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
In accordance with current European and national law, the data used in this study is only available for the researchers participating in this study. Thus, we are not allowed to distribute or make publicly available the data to other parties. However, researchers from public institutions can request Spanish data from SIDIAP if they comply with certain requirements. Further information is available online (https://www.sidiap.org/index.php/menu-solicitudesen/application-proccedure) or by contacting SIDIAP ([email protected]). Similarly, the Norwegian data in this study were delivered by the Norwegian registry holders to the researchers at UiO as pseudonymized data files. Data from the individual registries are available upon request to the registry holders, provided legal and ethical approvals.




