Deep learning-assisted model-based off-resonance correction for non-Cartesian SWI
Abstract
Purpose
Patient-induced inhomogeneities in the static magnetic field cause distortions and blurring (off-resonance artifacts) during acquisitions with long readouts such as in SWI. Conventional versatile correction methods based on extended Fourier models are too slow for clinical practice in computationally demanding cases such as 3D high-resolution non-Cartesian multi-coil acquisitions.
Theory
Most reconstruction methods can be accelerated when performing off-resonance correction by reducing the number of iterations, compressed coils, and correction components. Recent state-of-the-art unrolled deep learning architectures could help but are generally not adapted to corrupted measurements as they rely on the standard Fourier operator in the data consistency term. The combination of correction models and neural networks is therefore necessary to reduce reconstruction times.
Methods
Hybrid pipelines using UNets were trained stack-by-stack over 99 SWI 3D SPARKLING 20-fold accelerated acquisitions at 0.6 mm isotropic resolution using different off-resonance correction methods. Target images were obtained using slow model-based corrections based on self-estimated field maps. The proposed strategies, tested over 11 volumes, are compared to model-only and network-only pipelines.
Results
The proposed hybrid pipelines achieved scores competing with two to three times slower baseline methods, and neural networks were observed to contribute both as pre-conditioner and through inter-iteration memory by allowing more degrees of freedom over the model design.
Conclusion
A combination of model-based and network-based off-resonance correction was proposed to significantly accelerate conventional methods. Different promising synergies were observed between acceleration factors (iterations, coils, correction) and model/network that could be expanded in the future.
1 INTRODUCTION
Many parallel imaging and compressed-sensing (CS) methods1-7 have been proposed over the last two decades to accelerate MRI acquisitions. Non-Cartesian sampling patterns8, 9 have recently gained popularity through their capability to better exploit longer but fewer readouts. In particular, the Spreading Projection Algorithm for Rapid K-space sampLING (SPARKLING), proposed for 2D9 and 3D10, 11 imaging, responds to all degrees of freedom offered by modern MR scanners11 to fully explore k-space and match optimized target sampling densities. SWI,12 commonly used in high resolution brain venography or traumatic brain injuries,13 has been recently studied with SPARKLING11, 14 to reach acceleration factors (AF) higher than 15 in scan times compared to fully sampled Cartesian imaging in high resolution (0.6 mm) isotropic brain imaging. However non-Cartesian sampling patterns tend to be more sensitive to off-resonance artifacts causing geometric distortions and image blurring,15 notably with long readouts (e.g., 20 ms), thereby inducing k-space inconsistencies over the different gradient directions.16, 17 These artifacts emerge mostly from patient-induced static field inhomogeneities, notably pronounced near air-tissue interfaces, for instance in the vicinity of nasal cavity and ear canals.
Diverse methods have been proposed in the literature to correct those artifacts during the acquisition or image reconstruction. The spherical harmonic shimming technique is the current standard for all systems15, 18 but is generally limited to second or third-order harmonics, which already provide critical improvements. More advanced shim coil designs have been proposed recently19, 20 but still face technical and theoretical limitations.21 Post-processing methods can therefore be necessary as a complement for more demanding cases, such as Cartesian EPI22, 23 where alternating gradient direction at every time frame can be used to deduce and revert off-resonance induced geometric distortions. However, this technique is not applicable to non-Cartesian readouts (e.g., spirals,24-26 rosette,27 SPARKLING9-11) due to the multiple spatially-encoding gradients played simultaneously. Another less constraining and well-established method28-32 consists in compensating the undesired spatial variations by modifying the Fourier operator involved in image reconstruction in order to integrate prior knowledge on a field map. This technique can be applied to any imaging setup but considerably slows down (e.g., 15-fold) the image reconstruction process. The mandatory field map is directly available for multi-echo acquisitions,15 but it necessitates to be either externally collected by extending the scan time, or estimated.14, 33-38
Non-Cartesian acquisition strategies enable shorter scan times at the cost of increased image reconstruction duration. However, taking into account off-resonance correction within extended forward and adjoint operators has a multiplicative effect that makes the processing excessively long. In the recent years, deep learning (DL) has emerged for MRI reconstruction as a means to allow for improved image quality and faster processing, by similarly pushing the computation cost to offline training sessions. However, state-of-the-art network architectures are mostly focused on undersampling artifacts39-43 as they enforce data consistency with Fourier operators, which is inaccurate when dealing with off-resonance effects. More targeted literature invests considerable effort into estimating the field map,44, 45 already available in the context of SWI acquisitions (see Ref. 14 for details).
In this work, we study different approaches29-31 to model compressed representations of the non-Fourier operator involved in the data consistency term, and compensate for them using neural networks. The proposed extended non-Cartesian Primal-Dual network (NC-PDNet) architectures43 are trained to reproduce self-corrected14 high resolution SWI volumes based on highly accelerated multi-coil 3D SPARKLING11 trajectories (AF > 17) at 3T and each obtained through 8 h long reconstructions. This approximation allows us to reach a significant acceleration for model inversion with respect to three key elements, namely the number of unrolled iterations, compressed coils and correction components (i.e., interpolators involved in the non-Fourier operator), all contributing multiplicatively to the reconstruction time. The results are then compared to both model-only (CS reconstruction with non-Fourier operator) and network-only (original NC-PDNet) pipelines over 11 dedicated volumes and further decomposed to analyze the contributions of both neural networks and partially correcting models with respect to the three sources of approximation using tailored off-resonance metrics. The various benefits of hybrid architectures are demonstrated, with observed synergies paving the way to more improvements on image quality.
2 THEORY
2.1 Image reconstruction
For convenience we define the total number of samples (with the number of spokes and the number of samples per spoke) measured over the k-space and the total number of voxels (with , , and the image dimension in voxels).
2.2 Signal correction
For all coefficients, the computational load can be considerably reduced by taking advantage of the spoke redundancy (i.e., using the same decomposition over the spokes) and using histograms of the field map to solve a weighted version of Eq. (10), typically decreasing the image dimensions voxels to bins (see details in31). This way, the matrix is reduced from to and therefore correction coefficients can be obtained in a few seconds for high resolution 3D volumes.
We obtain from Eq. (9) a pseudo-Fourier operator (with representing the interpolation processing) that can be implemented as a wrapper for any regular Fourier operator and directly integrated into Eqs. (1) and (3). The same remark also holds for the adjoint Fourier operator .
2.3 Accelerated reconstruction and correction
The above mentioned correction technique is convenient but still increases the computation cost by multiplying factors of an already time-consuming reconstruction in the case of 3D high-resolution non-Cartesian imaging. Starting from the algorithm presented in Eqs. (4) and (5) combined with the correction operator in Eq. (9), different ways to reduce the computational burden can be explored by decreasing: The number of proximal gradient iterations , the number of channels , or the number of correction components . All possibilities were considered, and the last two are further explained in this subsection.
Different coil compression methods exist to decrease Q,53-55 but the most efficient ones, such as geometric coil compression,55 often exploit constraining k-space trajectory properties. More recent learning-based techniques have not been explored but may be considered in the future. Meanwhile, we used the trajectory-independent method by Buehrer et al.53 also based on SVD as it allows an efficient reduction of while also ordering compressed channels by explained variance, as represented in Figure 1 and detailed afterward. Some relations are observed between field maps and compressed channel sensitivities in the (Figure S7) but without strong demarcation; therefore, only the first components are kept.
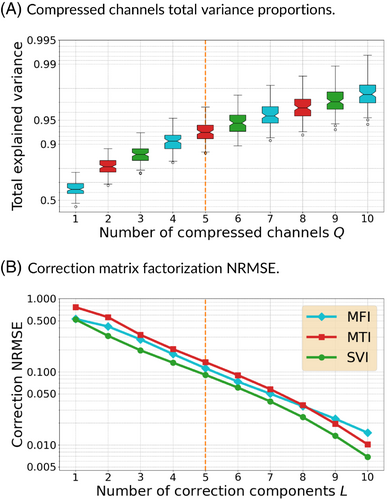
On the contrary, the SVI correction coefficients cover specific regions of the off-resonance spectrum (Figure S6). Simply using the first components, also studied hereafter, would mostly shift the data consistency focus toward low off-resonance areas that cover a much broader part of the brain images. A possibility to extend the spectrum coverage is to change the components used for data consistency over consecutive iterations. The SVI coefficients are more convenient for this, as two sets of and components with will share the same first components because of the orthogonality of the decomposition. This is not true for MFI and MTI methods as observed in Figures S4 and S5. It ensures that, while the first components carry the maximal amount of information, they are not redundant with the other components. Diverse strategies have been considered. The best performing solution updates data consistency between the first components and the following to components over iterations, called hereafter, to enforce fidelity toward either low or high off-resonance areas, respectively, in an alternated manner.
The goal is to provide improved reconstruction and correction while minimizing the processing time. , , and linearly multiply the time with exponentially decreasing quality contributions, as shown in Figure 1 and in supplementary materials with Table S4. Our approach is therefore to reduce the reconstruction load and recover the image quality using neural networks over compressed information.
3 METHODS
3.1 Proposed pipelines
Following the recommendations from Ramzi et al.,42 the end-to-end NC-PDNet architecture43 has been used as we consider 3D non-Cartesian SWI data. Particularly, we investigated the primal-only version where only the image domain processing is learned with an arbitrary neural network whereas data consistency is applied in k-space. However, in the context of 3D high resolution non-Cartesian and multi-coil imaging, the amount of graphics processing unit (GPU) memory required for a complete training is excessive. All or part of that memory can be moved to central processing unit CPU, but would result in a considerably longer training duration.
To address this issue, a solution often called greedy learning56 consists in breaking down the learning process into stacks trained sequentially to avoid any restriction on the number of iterations or the network size. An ideal case would be to have stacks consisting of at least one network pass followed by a data consistency block to still learn complementary features; however, the memory requirement increases drastically at the transition between single channel image domain and multi-coil k-space data. Therefore, the simplest proposition is to exclude the data consistency from the gradient computation by training an image-to-image network as represented in Figure 2, and then only apply data consistency and repeat for each stack.
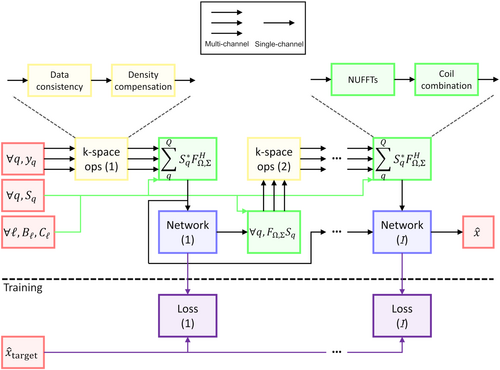
This modification requires adapting the feature originally named “memory”40 or more recently “buffer”,43 where networks could carry additional channels of information between iterations, as the additional network output channels are not all included in the backpropagation graph anymore. Instead, the network output is a single complex-valued volume but that will still be concatenated into a buffer of volumes from previous stacks to be used as input for the next stack. That way, the overall pipeline keeps the expressiveness required to learn CS-like acceleration schemes as originally suggested.40 The main drawback compared to end-to-end training is enforcing that each stack should yield the target results rather than letting each of the independent networks build intermediate states. However, this modification allows us to train architectures with an arbitrarily high number of coils and iterations.
The different partial correction strategies based on discussed in Section 2.3 are studied within an unrolled architecture in combination with UNets in the image domain. Indeed, conventional data consistency would either enforce the off-resonance artifacts in spite of the networks, or at best would not contribute to their correction. For all of the following studies, the variables , and have been retained to allow for 3D 0.6 mm isotropic corrections within 7–8 min of computing time at inference. The MFI, MTI and SVI coefficients are compared, along with the strategy, with buffers and pre-computed density compensations57 for the NUFFT operator from the gpuNUFFT1,58 and pysap-mri2,59 packages. The residual UNets are composed of three scales, each made of convolutional blocks of three layers with kernel size of and each followed by a ReLU activation (except for the final layer). The number of filters is doubled at each 3D downscale, with 16 filters at the first scale, and vice-versa for upscales. The complex nature of the input and output volumes is handled by considering real and imaginary parts as two separate real-valued channels. All deep-learning pipelines are composed of independent UNets, each with 390 066 parameters with buffers and 388 338 without. They are trained for 300 epochs, resulting in 100 h long trainings for each pipeline overall, with RAdam optimizer and learning rate of . The minimized cost function is the sum of an L1 loss applied to complex-valued images and multiscale SSIM over the magnitude images. All training experiments were run on the Jean-Zay supercomputer over a single NVIDIA Tesla V100 GPU with 32GB of VRAM.
3.2 Dataset
A total of 123 SWI volumes were acquired on patients with non-Cartesian 3D GRE sequences at 3T (Magnetom Prisma, Siemens Healthcare, Erlangen, Germany) with a 64-channel head/neck coil array. The protocol was approved by local and national ethical committees (IRB: CRM-2111-207). Patient demographics, a study flow diagram and an illustration of SPARKLING trajectories are provided in the supplementary materials (Section S1). The dataset covers a wide range of pathologies (aneurysm, sickle cell anemia, multiple sclerosis) and off-resonance related artifact levels.
The field maps were not acquired in order to avoid prolonging the exams. Instead self-estimated field maps were computed a posteriori using a recently published technique.14 To generate the ground truths, the field maps were used for model-based correction using the method described in31 with the SVI coefficients, resulting in approximately 8 h long reconstructions/corrections with , and over a single NVIDIA Tesla V100 GPU.
The dataset was then split into training (n = 99), validation (n = 11) and testing (n = 11) sets according to balanced age, gender, weight, off-resonance pre and post-correction visibility, and pathology type and visibility distributions. Two acquisitions were excluded due to strong motion (n = 1) and insufficient off-resonance correction (n = 1) caused by braces. In both cases, the self-estimated off-resonance correction still considerably improved the images. Minor quality concerns were raised regarding skin fat artifacts (n = 6) and partially deactivated readout coils (n = 2) without causing exclusion.
Although the trainings were carried out from complex-valued to complex-valued volumes, the SWI specific processing was applied afterwards for visualization and scoring as described in.12 The low frequencies were extracted by applying a Hanning window over the central third of k-space, before being removed from the phase image to obtain a high frequency map, subsequently normalized to produce a continuous mask. The magnitude image was multiplied five times by the mask, and a minimum-intensity projection (mIP) was computed using a thickness of 8 mm. All post-processing steps were again run on the Jean-Zay supercomputer.
3.3 Baselines
Diverse baseline models are proposed to assess the contributions of the different features involved in our pipeline. A first baseline detailed in the supplementary materials (Section S3) is given by replacing the neural networks by conventional CS reconstruction using sparsity promoting regularization in the wavelet domain (Symlet eight basis decomposed over three scales). The -norm was used for function with for thresholding the wavelet coefficients. To compensate the absence of buffers to learn acceleration schemes over iterations, the FISTA47 algorithm available in the pysap-mri package was used. The data consistency is implemented with with SVI coefficients reduced to . This baseline noted is used to determine to what extent the contribution of neural networks is critical for improved image quality.
The second baseline similarly replaces the correcting operator with the regular NUFFT while applying the same network-based regularization as the proposed pipeline.
Finally, the SVI and pipelines were tested without buffers, along with the baseline. The goal was to assess that the buffer feature, modified to fit the stacked-training setup, was still relevant even without end-to-end training.
3.4 Evaluation
4 RESULTS
The different pipelines are compared to each other hereafter, along with an ablation study of the modified buffer feature. Additional resources are provided in the supplementary materials, and notably more exhaustive CS baselines to quantify and understand the specific contributions of varying parameters , , and (Section S3). The number of coils in particular, as in Figure 3, is shown to have little influence on off-resonance correction, and the coil sensitivity profiles normalized in Eq. (3) seem to have more impact on all metrics than information carried by additional coils. In contrast, increasing contributes solely to off-resonance artifacts correction while augmenting positively impacts both off-resonance and high frequency content.
4.1 Correction pipelines
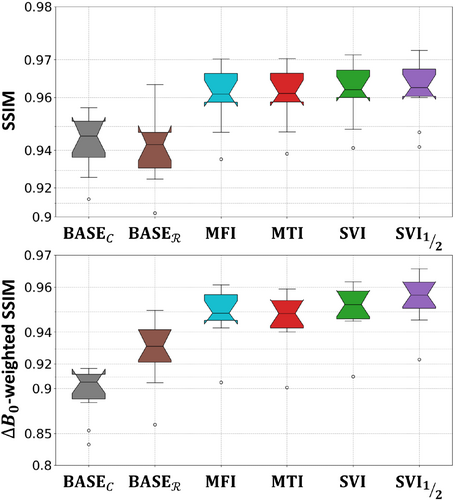
The different scores are reported in Table 1 and Figure 4, with corresponding pairwise -values in Table 2, and illustrated in Figure 5. Firstly, the baselines BASE and BASE in Table 1 demonstrate the expected positive contribution of a partially corrected Fourier operator against a basic model (BASE), and that of stacked UNets against wavelet-based regularization (BASE). The only statistically significant difference holds for -weighted SSIM, with a large improvement for BASE. Therefore, the proposed UNet architecture, when combined with conventional operator , is not capable of correcting off-resonance effects as much as a partially correcting operator does.
| Classic | -weighted | |||||
|---|---|---|---|---|---|---|
| Pipeline | Data consistency | Regularization | SSIM | PSNR | SSIM | PSNR |
| UNet | 0.9421 | 33.32 | 0.8947 | 23.50 | ||
| BASE | (SVI) | Wavelet | 0.9392 | 31.32 | 0.9249 | 23.33 |
| MFI | (MFI) | UNet | 0.9596 | 34.96 | 0.9475 | 26.42 |
| MTI | (MTI) | UNet | 0.9601 | 35.13 | 0.9452 | 26.11 |
| SVI | (SVI) | UNet | 0.9613 | 35.24 | 0.9498 | 26.57 |
| (SVI) | UNet | 0.9616 | 35.19 | 0.9541 | 27.31 | |
- Note: Various pipelines with , and are summarized with their data consistency and regularization terms and evaluated using classic and -weighted SSIM and PSNR scores.Bold signifies best (i.e. highest) PSNR and SSIM score.
| (a) Classic SSIM | ||||||
|---|---|---|---|---|---|---|
| BASE | MFI | MTI | SVI | |||
| - | ** | ** | ** | ** | ||
| BASE | - | ** | ** | ** | ** | |
| MFI | ** | ** | - | ** | ** | |
| MTI | ** | ** | - | ** | ** | |
| SVI | ** | ** | ** | ** | - | |
| ** | ** | ** | ** | - | ||
| (a) -weighted SSIM | ||||||
|---|---|---|---|---|---|---|
| BASE | MFI | MTI | SVI | |||
| ** | ** | ** | ** | ** | ||
| BASE | ** | ** | ** | ** | ** | |
| MFI | ** | ** | * | ** | ** | |
| MTI | ** | ** | * | ** | ** | |
| SVI | ** | ** | ** | ** | ** | |
| ** | ** | ** | ** | ** | ||
- Note: Various pipelines with , and are summarized with their data consistency and regularization terms and evaluated using classic and -weighted SSIM and PSNR scores.
-
p > 0.05
 .
. -
p < 0.05
 .
. -
p > 0.005
 .
.
Secondly, the different correction approaches (MFI, MTI, SVI) are combined with UNets through data consistency terms. All of them show significantly higher scores than both baselines. Their -weighted SSIM score follow the same ranking as those previously observed in Figure 1B prior to image reconstruction when solving Eq. (10): the SVI coefficients reach a better score as compared to MFI (second) and MTI (third) coefficients. This suggests that none of the diverse correction approaches carried by the different coefficients are better suited than another to help UNets compensate for the missing correction.
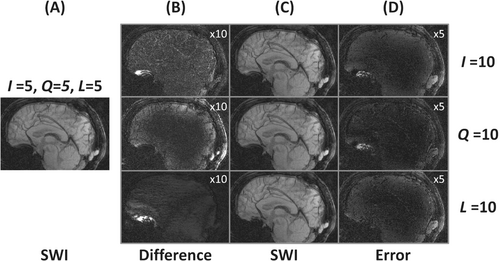
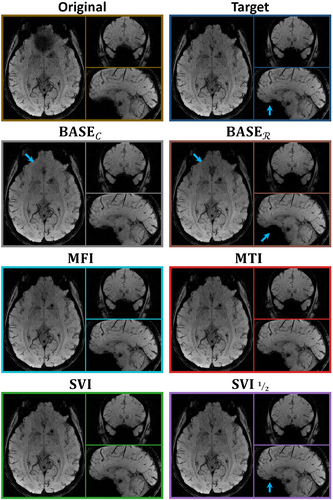
Finally, the pipeline improves the SVI approach with regard to -weighted SSIM, while not significantly deviating from it for classic SSIM. Exploring more components appears to guide the correction that networks alone could not achieve. This can be observed in Figure 5 near the bucco-nasal region visible with the sagittal views, where only recovers high-frequency details consistent with the target. The additional baselines from supplementary materials (Section S3) show that the pipeline competes with reconstruction two to three times slower depending on the evaluation criteria.
4.2 Network and model contributions
The reconstruction steps are further decomposed between data consistency and regularization in Figure 6 and illustrated in Figure 7. For both metrics, the two baselines shown in Figure 6 with brown (BASE) and gray (BASE) curves are again significantly different. The BASE curves show a slow start with oscillations between improving data consistency and degrading regularization but with a regular progression afterwards. The oscillations are explained by the constant wavelet threshold, too large during early iterations when the overall magnitude was still too low. In contrast, the networks used in BASE considerably improve the initialization, then followed by oscillations caused by the data consistency. Those oscillations can be explained by the off-resonance artifacts enforced through the wrong Fourier model but also by the sensitivity profiles altered by the normalization from Eq. (3) when reducing the number of channels . Both are illustrated in Figure 7 with green and blue arrows respectively.
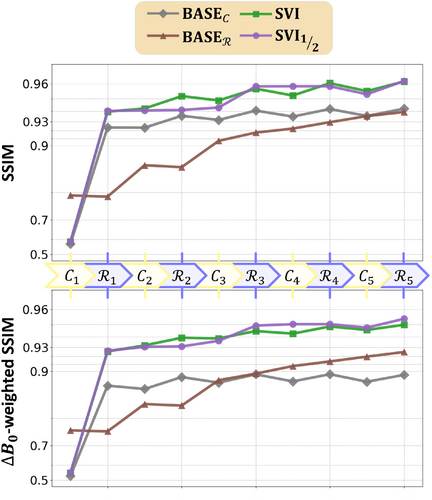
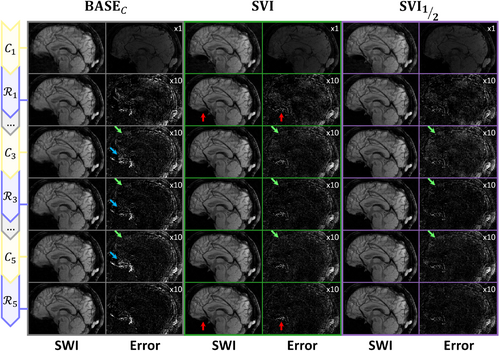
The proposed pipelines SVI and are observed to combine a steeper initialization and better progression over iterations, but still with sensitivity-related oscillations. The is however more robust to those as it alternates between large regions with heterogeneous sensitivities and smaller -specific regions. Overall, the UNets appear to contribute to all three studied factors, but the striking difference between BASE, SVI, and pipelines suggests that networks only recover low image frequencies when correcting off-resonance. The partial correction model is therefore required in those regions, but the networks serve as an effective image pre-conditioner.
4.3 Buffer feature ablation
The main pipelines have been evaluated with and without the proposed buffer feature modified to account for stacked training, namely SVI and along with BASE. The different results are shown in Table 3.
| Classic | -weighted | ||||||
|---|---|---|---|---|---|---|---|
| Pipeline | Buffers | p | SSIM | PSNR | p | SSIM | PSNR |
| - | - | 0.9427 | 33.40 | ** | 0.8979 | 23.69 | |
| 3 | 0.9421 | 33.32 | 0.8947 | 23.50 | |||
| SVI | - | * | 0.9603 | 35.11 | * | 0.9490 | 26.57 |
| 3 | 0.9613 | 35.24 | 0.9498 | 26.57 | |||
| - | ** | 0.9575 | 34.71 | ** | 0.9493 | 26.98 | |
| 3 | 0.9616 | 35.19 | 0.9541 | 27.31 | |||
- Note: The best performing pipelines (SVI and ) are evaluated with and without buffers, along with the baseline based on the non-correcting Fourier operator . The classic and -weighted scores are averaged over the testing dataset, with statistical significance assessed through pairwise two-sided Wilcoxon signed-rank tests over the SSIM scores with a global Benjamini-Hochberg correction.
-
p > 0.05
 .
. -
p < 0.05
 .
. -
p > 0.005
 .
.
The baseline BASE that combines UNets with conventional Fourier data consistency shows scores significantly worse when using the buffer feature. One interpretation could be that as consistency with corrupted brings back artifacts, as observed in Figure 7, adding memory of previous iterations only leads to a noisier learning process without additional information.
On the other hand, the approach shows significant and large improvements with buffers, more than for the SVI pipeline. However, it should be noted that classic scores without buffers are much lower than any other proposed pipeline, but still better than the baselines. This suggests that the exploratory correction strategy inherently degrades the overall reconstruction to advantage the off-resonance areas, but that buffer feature allows for a compensatory effect within neural networks.
5 DISCUSSION
The diverse contributions of this article are discussed hereafter. First, we compared conventional methods29-31 to perform, to the best of our knowledge, the first partial off-resonance correction study. Second, we analyzed the deep learning contributions in a multi-parametric acceleration pipeline. Finally, we conducted a deep learning study over a fairly large in vivo dataset consisting of model-based self-estimated off-resonance corrected volumes.14
5.1 Network and model improvements
The network and model contributions when facing reduced number of iterations, compressed coils, and correction components have been studied in Section 4.
Overall, the number of coils was observed to impact metrics by changing the sensitivity distribution superficially rather than improving the signal. Other normalization61 or coil compression methods62 could be explored in the future to at least balance this scoring issue. The proposed UNets appeared to mostly improve the initialization overall and help progression through the buffer feature when applying the strategy otherwise less efficient. The latter shows how deep learning allows for more flexibility when designing hybrid algorithms. However, high-frequency details are still only recovered through model-based correction.
More elaborated strategies could be developed to distribute efficiently the correction components over iterations. Particularly, for in-out SPARKLING trajectories the MTI coefficients were shown to correlate with high/low image frequencies. A better balance between low frequencies covered by neural networks and high frequencies enforced by specifically selected correction components might produce more reliable results.
5.2 Dataset generation
The map estimation technique developed in a previous publication14 assumes the relationship between inhomogeneities and image phase to be dominant over other phase sources for large echo times (e.g., 20 ms and higher) at 3T. It allows for a simple estimate of the field map retrospectively with minimal error and no motion-related mismatch for SWI acquisitions and alike. It also avoids any assumptions on the trajectory and can, therefore, be used on any dataset matching the above-mentioned acquisition setup and providing either the raw k-space data or complex-valued MR images.
The self-estimation method has been carefully assessed during the early stages of the study, and reached our expectations. However, two other competing methods were also considered, the first by Lee et al.38 through simulation from a binary mask, and the second by Patzig et al.35 through non-convex optimization. The simulation method in particular has been implemented in Python but the required mask obtained from the artifacted images was suboptimal. The main advantage of those three self-estimation methods for the purpose of our proposed acceleration pipeline is to require strictly the same data. This implies that the pipeline could be efficiently applied within the clinical context and additionally improve over sessions using the same acquisitions processed through longer and dedicated procedures.
6 CONCLUSIONS
MR acquisitions based on non-Cartesian acquisitions tend to exploit longer but fewer complex readouts to reduce overall scan times but suffer more from inhomogeneities. Diverse methods exist to compensate for these artifacts, but the few non-constraining ones slow down reconstructions by an unacceptably large factor (10–20 at 3T). Deep learning techniques have been developed in the recent years, but mostly to estimate the field map which can already be self-estimated using efficient models for SWI, or to compensate for undersampling artifacts in the CS setting. The proposed approach combines partial models and deep learning to ensure data fidelity at a low computation cost when addressing off-resonance artifacts and gain in image quality. It outlines both their individual and joint contributions. The MR volumes reconstructed at inference in only 7–8 min are competing with baselines obtained in 30 min to approximate 8 h long computations, and could be further accelerated by developing new hybrid strategies. Future work might also explore the parallel aspects of the correcting models to better fit memory constraints of the GPUs, or simply include undersampling compensation through self-supervised learning methods.
ACKNOWLEDGMENTS
The concepts and information presented in this article are based on research results that are not commercially available. Future availability cannot be guaranteed. This work was granted access to the HPC resources of IDRIS under the allocation 2021-AD011011153 made by GENCI. We thank Dr. Zaccharie Ramzi and Chaithya G R for their previous works and open-source contributions. Additionally we thank Cecilia Garrec for the scientifc editing in English. Philippe Ciuciu received funding from Siemens Healthineers (France) to support this research.