Quantification of MR spectra by deep learning in an idealized setting: Investigation of forms of input, network architectures, optimization by ensembles of networks, and training bias
Click here for author-reader discussions
Funding information: H2020 Marie Skłodowska-Curie Actions, Grant/Award Number: 813120; Nvidia, Swiss National Science Foundation, Grant/Award Number: 320030-175984
Abstract
Purpose
The aims of this work are (1) to explore deep learning (DL) architectures, spectroscopic input types, and learning designs toward optimal quantification in MR spectroscopy of simulated pathological spectra; and (2) to demonstrate accuracy and precision of DL predictions in view of inherent bias toward the training distribution.
Methods
Simulated 1D spectra and 2D spectrograms that mimic an extensive range of pathological in vivo conditions are used to train and test 24 different DL architectures. Active learning through altered training and testing data distributions is probed to optimize quantification performance. Ensembles of networks are explored to improve DL robustness and reduce the variance of estimates. A set of scores compares performances of DL predictions and traditional model fitting (MF).
Results
Ensembles of heterogeneous networks that combine 1D frequency-domain and 2D time-frequency domain spectrograms as input perform best. Dataset augmentation with active learning can improve performance, but gains are limited. MF is more accurate, although DL appears to be more precise at low SNR. However, this overall improved precision originates from a strong bias for cases with high uncertainty toward the dataset the network has been trained with, tending toward its average value.
Conclusion
MF mostly performs better compared to the faster DL approach. Potential intrinsic biases on training sets are dangerous in a clinical context that requires the algorithm to be unbiased to outliers (i.e., pathological data). Active learning and ensemble of networks are good strategies to improve prediction performances. However, data quality (sufficient SNR) has proven as a bottleneck for adequate unbiased performance—like in the case of MF.
1 INTRODUCTION
MR Spectroscopy (MRS) provides a noninvasive means for extracting biochemical profiles from in vivo tissues. Metabolites are encoded with different resonance frequency patterns, and their concentrations are directly proportional to the signal amplitude.1, 2 Metabolite quantification is traditionally based on model fitting (MF), where a parameterized model function is optimized to explain the data via a minimization algorithm. Metabolite parameters are usually estimated by a nonlinear least-squares fit (either in time or frequency domain) using a known basis set of the metabolite signals.3 However, despite various proposed fitting methods,3-7 robust, reliable, and accurate quantification of metabolite concentrations remains challenging.8 The major problems influencing the quantitative outcome are: (1) overlapping spectral patterns of metabolites, (2) low SNR, and (3) unknown background signals and line shape (no exact prior knowledge). Therefore, the problem is ill-posed, and current methods address it with different regularizations and constraint strategies (e.g., parameter bounds, penalizations, choice of the algorithm), with discrepancies in the results from one method to another.9
Supervised deep learning (DL) utilizes neural networks to discover essential features embedded in large data sets and to determine complex nonlinear mappings between inputs and outputs.10 Thus, DL does not require any prior knowledge or traditional assumptions. Given the success of the method in different areas,10-14 DL has been introduced into MRS as an alternative to conventional methods.15-22 Quantification of MRS datasets has been explored as follows: (1) DL algorithms identify datasets' features and either help reduce the parameter space dimension or set reliable starting conditions for the fit (i.e., combining knowledge on the physics with DL). It showed rapid spectral fitting of a whole-brain MRSI datasets.23 (2) Convolutional neural networks (CNNs) have been deployed to investigate combinations of spectral input of edited human brain MRS, which showed improved accuracy of straight metabolite quantitation when compared to traditional MF techniques.24 (3) Regression CNNs have been used to mine the real part of rat brain spectra to predict highly resolved metabolite basis set spectra with intensities proportional to the concentrations of the contributions,17 with results comparable to traditional MF approaches and showing readiness for (pre)clinical applications.22 (4) Targeting localized correlated spectroscopy (L-COSY) datasets, DL algorithms have reported faster data reconstruction and quantification compared to alternative acceleration techniques.16
Nevertheless, despite the reported equivalence in quantitation performance compared to traditional MF,14, 17, 22, 23 questions arise concerning the robustness of DL algorithms. A robust use within a clinical MRS context requires the algorithm to be unbiased also for pathological spectra. In imaging, DL has shown excellent performance for classification or segmentation tasks but may suffer from inherent weaknesses in subsets of representative outlier samples.11, 25 DL architectures for MRS quantitation have mostly been investigated for sample distributions of near-healthy spectral metabolite content. Hence, it can be suspected that high accuracy and precision are mainly found when DL is deployed for new entries of similar near-normal types. However, inaccurate estimates may result for tests with atypical datasets.26 Here, strongly variable metabolite concentrations that vary uniformly and independently over the entire plausible parameter space are used in the training set. This mimics the full range from healthy to strongly pathological spectra, that is, the full complexity of a clinical setup.
MRS signals are acquired in time domain but viewed in frequency domain. Traditional MF works in either of the two equivalent domains, and fit packages may allow the user to switch from one to the other for fitting and viewing. However, DL architectures for MRS quantification have mainly explored the frequency domain, mostly motivated by the reduced overlap between the constituting metabolite signals. Spectrograms18 present an extension into a simultaneous time/frequency domain representation and offer a 2D signal support that matches the input format for the original usage of CNN algorithms in computer vision. This work introduces a dedicated high-resolution spectrogram calculation focusing on signal-rich areas in both domains to be used as input for different CNN architectures. They are compared to other inputs and networks, inspired by previous MRS publications. Specifically, 24 network designs are investigated with differing input–output dataset types with a combined focus on depth (i.e., number of layers) and width (i.e., number of nodes/kernels) of the networks. This focus was motivated by the fact that the exploitation of spectrograms in deep learning has shown top-notch performance for speech and audio processing when deploying architectures with few layers and large convolutional kernels.27-29 Moreover, wide and shallow networks are more suitable to detect simple and small but fine-grained features. In addition, they are easier and faster to train.30 Network linearity (i.e., activation function) and locality (i.e., kernel size) are also investigated.
Besides investigating multiple architectures and input formats, two established main strategies for improving the outcome of predictions are also explored: active learning31 (data augmentation for critical types of spectra) and ensemble learning32, 33 (combination of outputs from multiple architectures).
Active learning can improve labeling efficiency,31, 34, 35 where the learning algorithm can interactively select a subset of examples that needs to be labeled. This is an iterative process where (1) the algorithm selects a subset of examples; (2) the subset is provided with labels; and (3) the learning method is updated with the new data.36 Uncertainty sampling37 is a specific strategy used in active learning that prioritizes selecting examples whose predictions are more uncertain (i.e., targeted data augmentation). Because these cases are usually close to the class separation boundaries, they contain most of the information needed to separate different classes.38, 39 In different applications, uncertainty sampling has been shown to improve the effectiveness of the labeling procedure significantly.34, 35, 37, 40
DL algorithms are sensitive to the specifics of the training.41 Hence, they usually find a different set of weights each time they are trained, producing different predictions.10 A successful approach for reducing the variance is to train multiple networks instead of one and combine their predictions.41 This is called ensemble learning, where the model generalization is maintained, but predictions improve compared to any of the single models.33 From a range of different techniques,42-44 here, stacking of models is implemented.32
To evaluate pros and cons of all these approaches, in silico ground truth (GT) knowledge is used (and hence no in vivo data was included in this evaluation) to assess performances via a dedicated set of metrics based on bias and SD. The CNN-predicted distributions of concentration are then compared to those from traditional MF. Furthermore, to emphasize the analysis at the core of the quantification task, the focus is placed on an idealized simulated setting with typical single-voxel spectra that have been preprocessed to eliminate phase as well as frequency drifts.3 This assumption aims at (1) freeing the MF algorithm from problems with local χ2 minima and (2) designing DL models optimized for the quantification task only.
2 METHODS
2.1 Simulations
This work is based on in silico simulations. A dataset of 22,500 entries was randomly split into 18,000 for training, 2000 for validation, and 2500 for testing. Larger dataset sizes are also explored, see section 2.4.
2.1.1 MR spectra
Brain spectra were simulated using actual RF pulse shapes for 16 metabolites at 3 T using Vespa45 for a semi-LASER46 protocol with TE = 35 ms, a sampling frequency of 4 kHz, and 4096 datapoints.
Further specifics of the simulations include: (1) Voigt line shapes, (2) metabolite concentration range, (3) addition of macromolecular background signal (MMBG), (4) noise generation, and (5) spectrum or spectrogram calculation.47 Metabolite concentrations vary independently and uniformly between 0 and twice a normal reference concentration for healthy human brain.1, 48-50 Maximal concentrations in mM units—NAA 25.8, tCr (1:1 sum of creatine + phosphocreatine spectra): 18.5, mI (myo-inositol): 14.7, Glu (glutamate): 20, Glc (glucose): 2, NAAG (N-acetylaspartylglutamate): 2.8, Gln (glutamine): 5.8, GSH (glutathione): 2, sI (syllo-inositol): 0.6, Gly (glycine): 2, Asp (aspartate): 3.5, PE (phosphoethanolamine): 3.3, Tau (taurine): 2, Lac (lactate): 1, and GABA (γ-aminobutyric acid): 1.8. The concentration for tCho (1:1 sum of glycerophosphorylcholine + phosphorylcholine spectra) ranges from 0 to 5 mM to mimic tumor conditions.51 A constant downscaled water reference (64.5 mM) is added at 0.5 ppm to ease quantitation. Metabolite T2s in ms (and hence Lorentzian broadening) are fixed to reference values from literature—tCr (CH2): 111, tCr (CH3): 169, NAA (CH3): 289, and all other protons: 185.49, 52-54 MMBG content, shim, and SNR mimicked in vivo acquisitions and varied independently and uniformly (time-domain water referenced SNR 5–40, Gaussian shim 2–5 Hz, MMBG amplitude ±33%). The MMBG pattern was simulated as a sum of overlapping Voigt lines as reported in Refs. 49 and 55 (Figure 1A).

2.1.2 Spectrograms
A spectrogram is a complex 2D representation of a spectrum, where frequencies vary with time: Every image column represents the frequency content of a particular time portion of the FID. Time information is binned along every row of the image. It is calculated via application of a short-time Fourier transform,18 where, depending on the size of the Fourier analysis window, different levels of frequency and time resolution can be achieved. A long window size modulated via zero-filling combined with a small overlap interval is chosen to increase frequency resolution and minimize the expense of time resolution (Figure 1B). Diagonal downsampling is designed to reduce the spectrogram size, keeping the original resolution grid at least as part of the time-frequency information on consecutive bins and reducing the spectrogram size (Figure 1C) to allow reasonable computation time for a CNN architecture (i.e., 128 frequency bins × 32 time bins) (Figure 1D).
2.2 Design and training of CNN architectures
A total of 24 different CNN architectures combined with different spectroscopic input representations are compared for MRS metabolite quantification. Current state-of-the art networks have been taken as reference models and adapted to the purpose and datasets used.
Scripts were written in Python56 using Keras library57 on a Tensorflow58 backend. Code ran on either of three graphic-processing units (GPUs; NVIDIA [Santa Clara, USA] Titan Xp, Titan RTX, or GeForce RTX 2080 Ti) or Google [Mountain View, USA] Colaboratory.59 Samples of the design are reported in Figure 2. Overall network designs are given in Table S1; Figures S1, S2, S3, S4, S5; and Text S1.
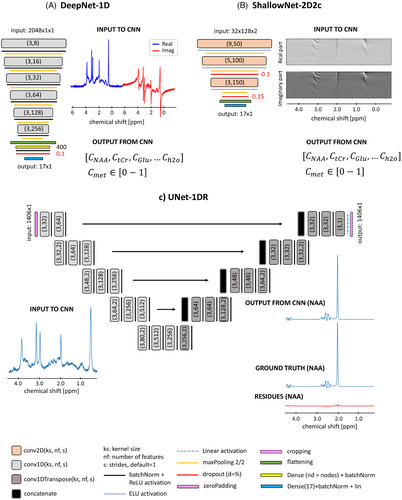
2.2.1 Architectures for straight numeric quantification of concentrations
A total of 22 architectures were fed with 1D (spectra) or 2D (spectrograms) input and mapped as output a vector of 17 normalized concentrations (i.e., in [0–1] interval) of 16 metabolites and the water reference, as listed in Table S1. Networks fed with 1D input exploit one channel with truncated spectra of 1024 datapoints from −0.5 to +6 ppm with concatenated real and imaginary parts (i.e., 2048 × 1 × 1 datapoints, Figure 2A). Networks fed with 2D input can either be configured in two channels (real and imaginary components of the spectrogram, 32 × 128 × 2 datapoints) or one channel (real and imaginary components concatenated, 64 × 128 × 1 datapoints, Figure 2B).
Five networks receive 1D input: two deep convolutional neural networks (DeepNet),60 two residual networks (ResNet)61 and one inception network (InceptionNet).62-64
This work investigates deep and shallow architectures either exploiting large or small convolutional kernel sizes. A total of 10 networks receive two-channel spectrograms as input. Given the limited size of the input FOV, the architecture is limited to be shallow (i.e., pooling operations to downsampling features directly following a convolutional layer are limited). However, a deeper architecture with multiple convolutional operations with sparse pooling is also compared. A further comparison is performed regarding the optimal activation function, comparing batch normalization + rectified linear unit (ReLU) versus exponential linear unit (ELU).65, 66 Seven networks receive one-channel spectrograms as input. With this configuration, deeper architectures are explored: two DeepNets, four ResNets, and one InceptionNet.
Architectures are analyzed either in a preconfigured parameter state or in a parameter space that had been optimized via Bayesian hyperparameterization.67 The optimization procedure is given in Text S1. In addition, to limit biases around zero for small concentrations,68 all network designs are characterized by a final layer with linear activation, allowing the prediction of negative concentrations.
2.2.2 Architectures for estimation of metabolite base spectra
- 1. UNet-1DR-hp : A total of 17 different networks with the same base architecture but adapted weights for each metabolite;
- 2. Unet-1DR-hp-met: A total of 17 different networks with adapted Bayesian-optimized architecture and weights for each metabolite.
Configurations are reported in Figure S5. First, metabolite concentrations are evaluated by feeding an input spectrum to the 17 metabolite-specific CNNs. Integration of the predicted metabolite base spectrum is then referenced to the integrated water reference to produce concentrations for a fully automated quantification pipeline.22
2.2.3 Training
Training and validation sets were randomly assigned for training the CNN on a maximum of 200 epochs with batch normalization of 50. The adaptive moment estimation algorithm (ADAM)70 was used with dedicated starting learning rates for each network.71, 72 The loss function was the mean-squared error (MSE). Visualization of training and validation loss over epochs combined with implementing an early-stopping criterion monitoring minimization of validation loss with patience = 10 has been used for tuning the network parameter space.57 Training time and test loss function are listed in Table S1.
2.2.4 Evaluation
- • a (slope of the regression line): must be close to 1 for ideal mapping of concentrations over the whole range of simulated metabolite content;
- • q (intercept of the regression line, mM): must be close to 0 to minimize prediction offsets/biases;
- • (coefficient of determination): must be close to 1 to assess full model explanation of the variability of the data;
- • (RMS error [RMSE] of prediction vs. GT, mM): as low as possible. However, expected to be comparable to Cramer Rao Lower Bounds (CRLBs) from MF.73
To easily compare different networks and input setups quantitatively in the Results section, these scores or combinations thereof have been used. The combinations are referred to as concise scores: as measure of linearity, to compare with CRLBs. q was excluded because it is mostly negligible.
2.3 Influence of inclusion of water reference peak
For the evaluation of the potential benefit of including a water reference peak, two slightly different ShallowNet-2D2c-hp networks are compared. Network A outputs 17 neurons (16 metabolites and water), whereas network B outputs 16 neurons only (no water output). Two adapted datasets are used for the investigation, one with (dataset A), and one without (dataset B) downscaled water reference at 0.5 ppm. Metabolite concentrations are calculated for both cases (assuming known water content in case A). Networks have been independently trained five times to monitor network variability over multiple trainings.
2.4 Active learning and dataset size
In this part, data augmentation techniques to smartly generate training sets are investigated. Subsets with 5000 new entries of the dataset where predictions scored worst are defined: specific subsets of spectrally weakly represented metabolites in either very low or very high concentrations and spectra with low SNR. New weighted datasets of 25,000 entries (20,000 training – 5000 validation set) or 40,000 entries (35,000 training – 5000 validation set) are generated (example in Figure 3, full description in Figure S6). Datasets with matching size and the testing set are kept unchanged from the previous simulation, thus with uniformly distributed concentrations and SNR. ShallowNet-2D2c-hp is selected as architecture and trained 10 times with a given augmented training set to minimize training variance.
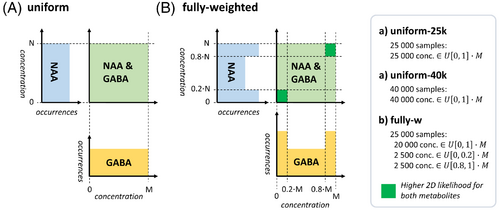
Complementarily, given the network trained on a uniform span of concentrations, active learning is investigated in the testing phase on three different test sets where concentrations are clipped to a progressively smaller range of 20%–80%, 20%–80% with SNR >20, and 40%–60% concentration range relative to the training set.
2.5 Ensemble of networks
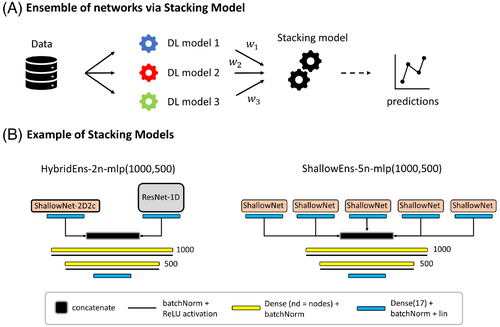
In this section, ensembles of networks are implemented via stacking of models.32 This consists of designing a DL architecture called stacking model (a multilayer perceptron (MLP) with two hidden layers is selected for this case) that will take as input the combination of a given number of independently pretrained models. The stacking model aims at weighting predictions from single models. It is trained using the same training and validation sets used to train single models while keeping the weights of the pretrained input models fixed. Three different ensembles are investigated: ShallowEns-5n groups five identical ShallowNet-2D2c-hp architectures, whereas HybridEns tests heterogeneous inputs grouping either two or 10 different networks (ShallowNet-2D2c-hp and ResNet-1D-hp) (Figure 4).
2.6 Model fitting
Spectra are fitted using FiTAID7 given its top performance in the ISMRM fitting challenge9 and to be expected for the spectra as used in the current setup (in particular, without undefined spurious baseline). The model consists of a linear combination of the metabolite base spectra with Voigt lineshape, where the Lorentzian component was kept fixed at the known GT value. The areas of the metabolites are restricted in a range corresponding to [], where μ is the average concentration in the testing set distribution (i.e., the normal tissue content). These bounds mimic the effective boundaries of the DL algorithms. CRLBs are used as a precision measure74 and are considered for three subgroups of the testing set (high [SNR > 28.4], medium [16.7 < SNR < 28.4], and low [SNR < 16.7] relative SNR, respectively).
3 RESULTS
3.1 S1Metabolite quantification referenced to the downscaled water peak
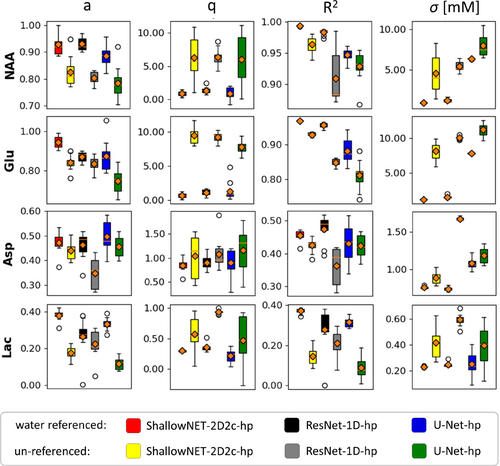
As illustrated for three different networks, Figure 5 shows that CNN predictions perform better if the spectra are referenced to a downscaled water peak: Regression slope a and are closer to 1; is appreciably lower. Moreover, the spread of the scores is on average reduced, displaying improved stability over multiple trainings. Extended results are presented in Figures S7 and S8.
3.2 Network design
Figure 6 reports CNN predictions versus GT values of a ResNet-1D-hp architecture for nine metabolites (see Figures S9 and S10 for extended results on 16 metabolites or different CNN architecture). Distributions of GT and predicted values are displayed for the test set (as for all results). Predictions relate very well to the GT for well-represented metabolites (top row). However, for metabolites with lower relative SNR, predicted distributions of concentrations tend to be less uniform and are biased toward average values of the GT distributions. Thus, concentrations at distribution boundaries are systematically mispredicted, particularly for low SNR. This is reflected in lower and values and higher σ. Figures S11 and S12 include a comparison of multiple networks via bar graphs (which are ill-suited to express the systematic bias) and a plot of distributions of predictions.
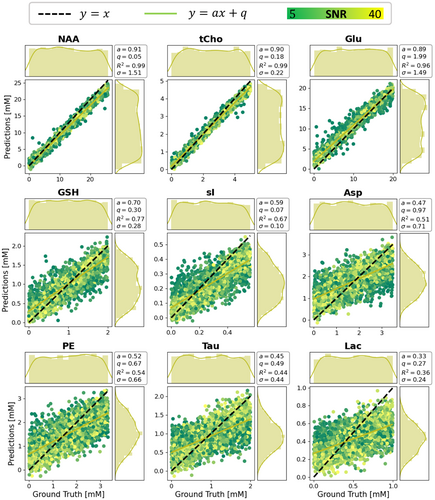
- 1. Well-represented metabolites: NAA, tCho, tCr, mI, Glu with averaged DL scores and < 15%, as well as MF scores and ;
- 2. Medium-represented metabolites: Glc, NAAG, Gln, GSH with averaged DL scores and , as well as MF scores and ;
- 3. Weakly represented metabolites: sI, Gly, Asp, PE, Tau, Lac, GABA with averaged DL scores and average , as well as MF scores and .
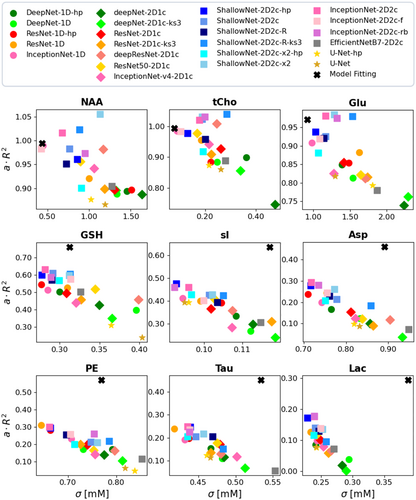
Overall, multiple DL networks perform similarly, but some general differences are noteworthy. Optimized spectrogram representation via two channels combined with a shallow architecture (i.e., dark blue squares) is found to be well suited for MRS quantification, showing mostly better performances than alternative deeper designs (i.e., light blue, pink, and gray squares), with one-channel designs (diamonds) or 1D spectra as signal representation (circles). Benefits are evident for medium and weakly represented metabolites. Performances of direct quantification and two-step quantification via base spectrum prediction followed by integration (stars) are similar. MF is found superior to DL for all medium- and weakly represented metabolites with significant average improvements for However, σ tends to be higher for many cases. A more detailed presentation of performance is given in Figures S14 and Text S2.
Figure 8 displays plots of prediction errors (i.e., Δ = prediction−GT) and their spread σ as a functionof SNR and shim for tCho, NAAG, and sI. Prediction uncertainties increase with noise level approximately linearly with 1/SNR and reach a plateau for weakly represented metabolites when the spread represents essentially the whole training range. No dependence on shim is apparent for the investigated range.

3.3 Dataset size, active learning, and ensembles of networks
Figure 9 reports on performance improvements by active learning in training phase and dataset sizing (part 9A) as well as by using an ensemble of networks (part 9B) for four metabolites as reflected by concise scores. Outcomes of emulated active learning approaches in limiting the testing sets are illustrated through regression plots for Gln in Figure 9C. Detailed comparisons for 16 metabolites are given in Figure S15, Table S2, Table S3, Figure S16, and Table S4.
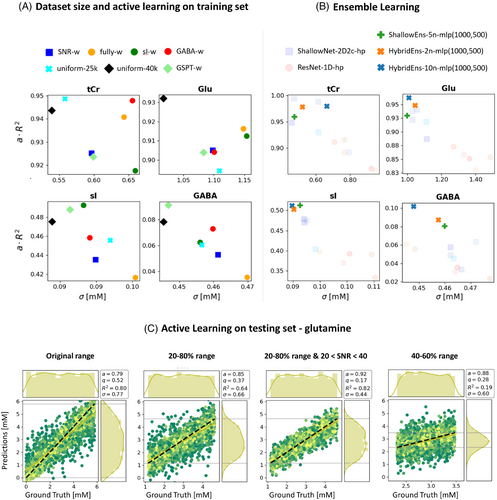
3.3.1 Dataset size
The performance showed moderate improvements for most metabolites when dataset size was increased from 25,000 to 40,000 samples (Figure S9).
3.3.2 Active learning
Dataset augmentation to favor training with combinations of low or high concentrations of weakly represented metabolites (see Figure S6B–S6D) does not substantially improve performance (Figure 9A, Figure S15, Table S2). Mild improvements (<6% for a, q, and ) are seen for GABA and sI, respectively, when exploiting metabolite-specifically augmented datasets (GABA-w, sI-w). Increased dataset size combined with data augmentation to favor high and low concentrations of different metabolites (GSPT-w) moderately improves performances for the augmented metabolites (GABA, sI, PE, Tau). It also extends mild improvements to medium- to weakly represented metabolites that have not undergone data augmentation (e.g., Lac, Gly, Gln). A dataset that is strongly weighted toward extreme combinations of low or high concentration for all metabolites (fully-w) or a dataset weighted toward low SNR (SNR-w) deteriorated performances.
Clipping the test set to 20%–80% or 40%–60% of the concentration range in training renders improved performances (on average a + 4.5%, q−10.2%, −23.9% and a + 4%, q−37.5%, −36.2%, respectively), which is even enhanced further when the testing set includes samples with higher SNR (on average a + 15.4%, q−45.4%, −36.2%). Given the limited range on the y-axis, is less representative (Figure 9C, Table S3).
3.3.3 Ensemble of networks
Ensembles of Bayesian-optimized networks show consistent and relevant improvements for medium- to weakly represented metabolites without deteriorating performance for well-defined metabolites. A hybrid ensemble outperforms the ensemble of networks of the same type. The performance of the ensemble increases with the number of combined networks (Figures 9B, S16) (Table S4).
3.4 CNN predictions versus model fitting estimates
A general juxtaposition of CNN and MF performance is contained in Figure 7. In Figure 10, detailed results are presented for two metabolites in the form of regression plots for ShallowNet-2D2c-hp and MF with FiTAID. In addition, the estimated CRLBs from MF are displayed and then compared in subgroups of SNR with the variance found in MF estimates and CNN predictions.
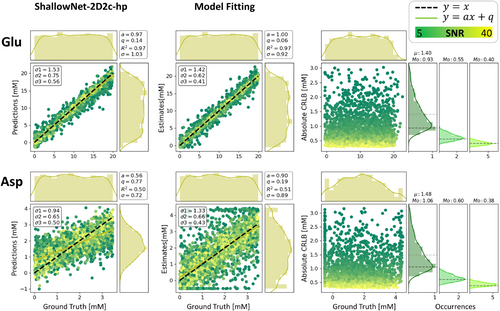
Area-constrained MF shows biases at the parameter boundaries for weakly represented metabolites (e.g., Asp). However, traditional MF outperforms quantification via DL: regression lines show less bias (a and q), and the distribution shape of estimates is closer to a uniform pattern within the GT range. RMSEs (σs) are higher in the case of MF for medium- to weakly represented metabolites (e.g., Asp) but lower for well-defined metabolites (e.g., Glu) (as formerly noted in Figure 7). Consequently, although σs of MF are bigger than the CRLBs estimated for their SNR reference group, σs of DL overestimate CRLBs for well-defined metabolites and underestimate CRLBs for weakly represented metabolites.
4 DISCUSSION
Quantitation of brain metabolites using deep learning methods with spectroscopy data in 1D, 2D, and a combined input format was implemented in multiple network architectures. The main aim of the investigation was to compare the core performance of quantification in an idealized setting of simulated spectra. In fact, the analysis of the optimal performance of both, MF and DL, may otherwise be blurred by additional experimental inaccuracies or artifacts from actual in vitro or in vivo spectra. Moreover, these nuisance contributors may be tackled in separate traditional or DL preprocessing steps that are beyond the current analysis. Many of the methods proved successful in providing absolute concentration values even when using a very large concentration range for the tested metabolites that goes way beyond the near-normal range that has often been used in the past. In addition, different forms of network input were tested, including a specifically tailored time-frequency domain representation and a downscaled water peak for easing of quantification. Whereas data augmentation by active learning schemes showed only modest improvements, ensembles of heterogeneous networks that combine both input representation domains improve the quantitation tasks substantially.
Results from DL predictions were compared to estimates from traditional MF, where it was found that MF is more accurate than DL at high and modest relative noise levels. MF yields higher variance at low SNR, with estimated concentrations artificially aggregated at the boundaries of the fitting parameter range. Predictions obtained with DL algorithms delusively appear more precise (lower RMSE) in the low SNR regime, which may misguide nonexperts to believe that the DL predictions are reliable even at low SNR. However, these predicted concentrations are strongly biased by the dataset the network has been trained with. Hence, in case of high uncertainty (e.g., metabolites with low relative SNR or present in concentrations at the edge of the parameter/training space), the predicted concentration tends toward the most likely value: the average value from the training set.
4.1 Forms of input to networks
Previously, 1D spectra have mostly been used as input for DL algorithms. Here, they have been compared and combined with 2D time-frequency domain spectrograms that had explicitly been designed to be of manageable size while retaining those areas of the high-resolved standard spectrogram that contain the most relevant information, that is, rich in detail in frequency domain to distinguish overlapping spectral features but also maintaining enough temporal structure to characterize signal decay. This comes at the cost that the spectrogram creation cannot be reversed mathematically. However, this is irrelevant when serving as input to a DL network. It was found that this tailored time-frequency representation as input in combination with a shallow CNN architecture performs best and outperforms the use of traditional 1D frequency-domain input for straight quantification or for metabolite basis spectrum isolation with subsequent integration. Furthermore, DL quantitation performance improved upon the inclusion of a downscaled water peak for reference, likely solving scaling issues if no reference is provided.
4.2 Active learning
Active learning has been explored by extending the training dataset with cases that appeared challenging to predict in the original setup. In particular, new training data with nonequal distribution of metabolite concentrations have been used with a predominance of single or multiple metabolites at low or high concentrations. None of these trials led to substantial improvements, although it might be helpful if specific metabolites are targeted primarily. Such data augmentation for all metabolites simultaneously even deteriorated the overall network performance. This can be understood given that augmentation at the border of the concentration range inherently leads to an underrepresentation of intermediate cases, which are equally relevant for the overall performance. Extending the size of the training set even further in an unspecific manner appears to still yield modest improvements.75 In addition, an unconventional way of active learning was probed by using unequal dataset ranges in training and testing by limiting testing on the central portion of the training range. This setup clearly ameliorated some of the issues at the edges of the testing range found in the typical setup. This approach was only implemented by reducing the test range rather than expanding the training range, which would yield better comparable outcome scores (e.g., R2). However, expanding the training range to negative concentrations may be questionable.
While data augmentation with a bigger proportion of low SNR spectra leads to worse performance, the theoretical prediction limits for good SNR data are probed in the noiseless scenario in which training and testing are run with GT data. Example results for a ShallowNet architecture are reported in Figure S17 for NAA, GSH, and Lac. This, combined with the results discussed, suggests that the bottleneck that limits higher prediction performances is SNR, just like in traditional MF, regardless of the implementation of state-of-the-art networks, network optimization, or dataset augmentation. It thus reflects limitations in clinical applications where high enough SNR is just not available. According to this study, DL cannot do miracles unless one accepts the bias toward training conditions.73
4.3 Ensemble of networks
An ensemble of networks has been implemented, and it shows improvements for quantifying metabolites. A combination of networks is less sensitive to the specifics of the training and helps reduce the variance in the predictions. Furthermore, ensembles of networks where multiple noise-sensitive predictions are weighted are more robust to noise. However, even the thus optimized networks underperform in comparison to MF. For MF, CRLBs clearly indicate limits for the confidence in the fit results. For DL, including the optimized ensemble of networks, such limits can only vaguely be deduced from the distributions of predicted values with the major danger of bias toward training data norms.76, 77 The CRLB would provide good guidance for the valid range of DL predictions as well—although of course they are not readily available without the model. New tools to estimate precision and replace CRLB in the case of DL76, 77 still have to prove their value in practice. The situation will be different again if the DL quantification is trained to include cleaning of spectra from artifacts (ghosts, baseline interference) where CRLBs are not available.
4.4 Low SNR regime
Both MF and DL show lower reliability in quantifying metabolites in the low SNR regime. Clear-cut SNR limits for validity of concentration estimates are not available, neither for MF nor for DL, although SNR values are often indicated as measure of spectral quality. While CRLBs provide a widely used and easy-to-interpret reliability measure that includes the influence of SNR, a similar widely accepted concept does currently not extend to DL approaches.77 Obviously, a SNR threshold for DL reliability would have to be metabolite-SNR specific, but already the definition of a meaningful metabolite-specific SNR would be cumbersome given that peak-splitting patterns and number of contributing protons as well as lineshape introduce ambiguity. On top, such a metabolite SNR would depend on the estimated metabolite content, whose reliability is at stake. Therefore, just like for MF, global or metabolite-specific SNR will not be informative enough. An uncertainty measure is needed that is based on the predictions and noise distribution but also integrating the uncertainty propagation of the DL model prediction78, 79 (like the inverse of the Fisher information matrix used in the CRLB definition74). Despite flourishing literature,80, 81 addressing uncertainty estimation as a complementary tool for DL interpretability, a full-scale analysis of the robustness and reliability of such models is still challenging.82-84 First attempts to extend these concepts in DL for MRS quantification are just subject of recent investigations76, 77 but far from general acceptance.
4.5 Limitations
The current investigation focused on probing multiple DL techniques and input forms for a full range of metabolite concentrations but a limited range of spectral quality. In particular, the shim remained in a broadly acceptable range, no phase or frequency jitter was considered, and no artifactual data was included. Such features could have been integrated in the current setup to arrive at a more realistic framework. However, the core of the findings (performance of the actual quantification step) is expected to remain in place. In addition, it is recommended to add separate preprocessing steps to prepare the data for the presented algorithms rather than to combine processing and quantification in a single process.3 They could be realized in the form of dedicated DL networks, such as those proposed for phase and frequency drift corrections,20, 85, 86 and stacked before the quantification model. This would also ensure the essential gain in speed expected from DL quantification models.
Direct comparison with previously proposed successful DL quantification implementations like Ref. 22 was not possible or meaningful for lack of open access network details and differences in the considered spectra.
Our particular implementation used to create spectrograms was optimized to maintain relevant resolution but downweights the initial part of the FID (initialization of Hamming window). CNN inputs may thus not be fully susceptible to changes in broad signals. Alternative recipes with, for example, prefilled filters or circular datasets, were not explored.
Furthermore, active learning has been explored for a single network type and could in principle be more beneficial for other networks or types of input than what has been found here.
5 CONCLUSIONS
Quantification of MR spectra via diverse and optimized DL algorithms and using 1D and 2D input formats have been explored and have shown adequate performance as long as the metabolite-specific SNR is sufficient. However, as soon as SNR becomes critical, CNN predictions are strongly biased to the training dataset structure.
Traditional MF requires parameter tuning and algorithm convergence, making it more time consuming than DL-based estimates. On the other hand, we have shown that ideally (i.e., with simulated cases) and statistically (i.e., within a variable cohort of cases), it can achieve higher performances when compared to a faster DL approach. DL does not require feature selection by the user, but the potential intrinsic biases at training set boundaries act like soft constraints in traditional modeling,9 leading estimated values to the average expected concentration range, which is dangerous in a clinical context that requires the algorithm to be unbiased to outliers (i.e., pathological data).
Active learning and ensemble of networks are attractive strategies to improve prediction performances. However, data quality (i.e., high SNR) has proven as bottleneck for adequate unbiased performance.
ACKNOWLEDGMENTS
This project has received funding from the European Union's Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement # 813120 (inspire-med) and the Swiss National Science Foundation (#320030-175984). We acknowledge the support of NVIDIA Corporation for the donation of a Titan Xp GPU used for some of this research. The authors thank Prof Maurico Reyes (ARTORG Center for Biomedical Engineering Research, University of Bern, Switzerland) for very helpful discussions.
Open Research
DATA AVAILABILITY STATEMENT
The main part of the code will be available on GitHub (https://github.com/bellarude). In addition, simulated datasets will be available on MRSHub (https://mrshub.org/). For questions, please contact the authors.




