Marmoset brain segmentation from deconvolved magnetic resonance images and estimated label maps
Corresponding Author
Farah Bazzi
Computer Science Research Institute of Toulouse (IRIT), Toulouse University UPS, CNRS, UMR, Toulouse, France
Centre de Recherche Cerveau et Cognition (CerCo), Université de Toulouse UPS, CNRS, UMR, Toulouse, France
Doctoral School of Sciences and Technology, AZM Center for Research in Biotechnology and Its Applications, Lebanese University, Beirut, Lebanon
Correspondence
Farah Bazzi, Institut de Recherche en Informatique de Toulouse (IRIT), Université de Toulouse UPS, CNRS, UMR 5505, Toulouse, France.
Email: [email protected]
Search for more papers by this authorMuriel Mescam
Centre de Recherche Cerveau et Cognition (CerCo), Université de Toulouse UPS, CNRS, UMR, Toulouse, France
Search for more papers by this authorAhmad Diab
Doctoral School of Sciences and Technology, AZM Center for Research in Biotechnology and Its Applications, Lebanese University, Beirut, Lebanon
Search for more papers by this authorOmar Falou
Doctoral School of Sciences and Technology, AZM Center for Research in Biotechnology and Its Applications, Lebanese University, Beirut, Lebanon
Search for more papers by this authorHassan Amoud
Doctoral School of Sciences and Technology, AZM Center for Research in Biotechnology and Its Applications, Lebanese University, Beirut, Lebanon
Search for more papers by this authorAdrian Basarab
Computer Science Research Institute of Toulouse (IRIT), Toulouse University UPS, CNRS, UMR, Toulouse, France
Search for more papers by this authorDenis Kouamé
Computer Science Research Institute of Toulouse (IRIT), Toulouse University UPS, CNRS, UMR, Toulouse, France
Search for more papers by this authorCorresponding Author
Farah Bazzi
Computer Science Research Institute of Toulouse (IRIT), Toulouse University UPS, CNRS, UMR, Toulouse, France
Centre de Recherche Cerveau et Cognition (CerCo), Université de Toulouse UPS, CNRS, UMR, Toulouse, France
Doctoral School of Sciences and Technology, AZM Center for Research in Biotechnology and Its Applications, Lebanese University, Beirut, Lebanon
Correspondence
Farah Bazzi, Institut de Recherche en Informatique de Toulouse (IRIT), Université de Toulouse UPS, CNRS, UMR 5505, Toulouse, France.
Email: [email protected]
Search for more papers by this authorMuriel Mescam
Centre de Recherche Cerveau et Cognition (CerCo), Université de Toulouse UPS, CNRS, UMR, Toulouse, France
Search for more papers by this authorAhmad Diab
Doctoral School of Sciences and Technology, AZM Center for Research in Biotechnology and Its Applications, Lebanese University, Beirut, Lebanon
Search for more papers by this authorOmar Falou
Doctoral School of Sciences and Technology, AZM Center for Research in Biotechnology and Its Applications, Lebanese University, Beirut, Lebanon
Search for more papers by this authorHassan Amoud
Doctoral School of Sciences and Technology, AZM Center for Research in Biotechnology and Its Applications, Lebanese University, Beirut, Lebanon
Search for more papers by this authorAdrian Basarab
Computer Science Research Institute of Toulouse (IRIT), Toulouse University UPS, CNRS, UMR, Toulouse, France
Search for more papers by this authorDenis Kouamé
Computer Science Research Institute of Toulouse (IRIT), Toulouse University UPS, CNRS, UMR, Toulouse, France
Search for more papers by this authorFunding information
The AZM and SAADE Association and the Doctoral School of Science and Technology, Lebanon
Abstract
Purpose
The proposed method aims to create label maps that can be used for the segmentation of animal brain MR images without the need of a brain template. This is achieved by performing a joint deconvolution and segmentation of the brain MR images.
Methods
It is based on modeling locally the image statistics using a generalized Gaussian distribution (GGD) and couples the deconvolved image and its corresponding labels map using the GGD-Potts model. Because of the complexity of the resulting Bayesian estimators of the unknown model parameters, a Gibbs sampler is used to generate samples following the desired posterior probability.
Results
The performance of the proposed algorithm is assessed on simulated and real MR images by the segmentation of enhanced marmoset brain images into its main compartments using the corresponding label maps created. Quantitative assessment showed that this method presents results that are comparable to those obtained with the classical method—registering the volumes to a brain template.
Conclusion
The proposed method of using labels as prior information for brain segmentation provides a similar or a slightly better performance compared with the classical reference method based on a dedicated template.
Supporting Information
| Filename | Description |
|---|---|
| mrm28881-sup-0001-Supinfo.pdfPDF document, 1.2 MB |
FIGURE S1 Hierarchical Bayesian model for the parameters' and hyperparameters' priors, where the deblurred image FIGURE S2 Showing the native axial slice with the manual GM and WM segmentation compared to the deblurred images and their respective segmentations with the noise variance being pre-estimated from the background signals, pre-estimated by a Rician noise estimator and modelled, respectively FIGURE S3 The effect of varying the value of the granularity coefficient FIGURE S4 SSIM index and ISNR computed between the deconvolved image and the ground truth for different MCMC iterations and the elapsed time (in secs) for each set of iterations to be completed FIGURE S5 Effect of varying the number of iterations on the estimated labels map FIGURE S6 Showing simulated GM, WM and CSF regions with MoG distributions that: (A) perfectly fits a GGD distribution, (B) have a good GGD fit and (C) have a bad GGD fit FIGURE S7 The simulated, corrupted and recovered images along with their estimated labels maps for the three cases where the regions were created with MoG distributions that: (A) perfectly fits a GGD distribution, (B) have a good GGD fit and (C) have a bad GGD fit FIGURE S8 Showing GM, WM and CSF segmentation of the images simulated with a perfect GGD fit, a good GGD fit and a bad GGD fit compared to the true labels FIGURE S9 An axial slice of the experimental results done on an additional real MR volume showing the segmentation results of GM, WM and CSF with segTemp and segLabels TABLE S1 Parameters of the Gaussian distributions forming the MoG of each region TABLE S2 SSIM index and DICE coefficients for GM, WM and CSF segmentation of the three simulated images compared to the true labels |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
REFERENCES
- 1Wang L, Chitiboi T, Meine H, Günther M, Hahn HK. Principles and methods for automatic and semi-automatic tissue segmentation in MRI data. Magn Reson Mater Phys, Biol Med. 2016; 29: 95-110.
- 2Schick F. Tissue segmentation: a crucial tool for quantitative MRI and visualization of anatomical structures. Magn Reson Mater Phy. 2016; 29: 89-93.
- 3Lorenzi M, Pennec X, Frisoni GB, Ayache N. Disentangling normal aging from Alzheimer’s disease in structural magnetic resonance images. Neurobiol Aging. 2015; 36: S42-S52.
- 4Baillard C, Hellier P, Barillot C. Segmentation of brain 3D MR images using level sets and dense registration. Med Image Anal. 2001; 5: 185-194.
- 5Kapur T, Grimson WEL, Wells WM, Kikinis R. Segmentation of brain tissue from magnetic resonance images. Med Image Anal. 1996; 1: 109-127.
- 6Gordillo N, Montseny E, Sobrevilla P. State of the art survey on MRI brain tumor segmentation. Magn Reson Imaging. 2013; 31: 1426-1438.
- 7Balafar MA, Ramli AR, Saripan MI, Mashohor S. Review of brain MRI image segmentation methods. Artif Intell Rev. 2010; 33: 261-274.
- 8Shattuck DW, Sandor-Leahy SR, Schaper KA, Rottenberg DA, Leahy RM. Magnetic resonance image tissue classification using a partial volume model. Neuroimage. 2001; 13: 856-876.
- 9Kamnitsas K, Ledig C, Newcombe VFJ, et al. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal. 2017; 36: 61-78.
- 10Amato U, Larobina M, Antoniadis A, Alfano B. Segmentation of magnetic resonance brain images through discriminant analysis. J Neurosci Methods. 2003; 131: 65-74.
- 11Anbeek P, Vincken KL, van Bochove GS, van Osch MJP, van der Grond J. Probabilistic segmentation of brain tissue in MR imaging. Neuroimage. 2005; 27: 795-804.
- 12Dong F, Peng J. Brain MR image segmentation based on local Gaussian mixture model and nonlocal spatial regularization. J Vis Commun Image Represent. 2014; 25: 827-839.
- 13Caldairou B, Passat N, Habas P, Studholme C, Rousseau F. A non-local fuzzy segmentation method: application to brain MRI. Pattern Recogn. 2011; 44: 1916-1927.
- 14Ballester MÁG, Zisserman A, Brady M. Segmentation and measurement of brain structures in MRI including confidence bounds. Med Image Anal. 2000; 4: 189-200.
- 15Feo R, Giove F. Towards an efficient segmentation of small rodents brain: A short critical review. J Neurosci Methods. 2019; 323: 82-89.
- 16Zhao G, Liu F, Oler JA, Meyerand ME, Kalin NH, Birn RM. Bayesian convolutional neural network based MRI brain extraction on nonhuman primates. NeuroImage. 2018; 175: 32-44.
- 17McLaren DG, Kosmatka KJ, Kastman EK, Bendlin BB, Johnson SC. Rhesus macaque brain morphometry: a methodological comparison of voxel-wise approaches. Methods. 2010; 50: 157-165.
- 18Ashburner J, Friston KJ. Unified segmentation. Neuroimage. 2005; 26: 839-851.
- 19Hikishima K, Ando K, Komaki Y, et al. Voxel-based morphometry of the marmoset brain: in vivo detection of volume loss in the substantia nigra of the MPTP-treated Parkinson’s disease model. Neuroscience. 2015; 300: 585-592.
- 20Hikishima K, Quallo MM, Komaki Y, et al. Population-averaged standard template brain atlas for the common marmoset (Callithrix jacchus). Neuroimage. 2011; 54: 2741-2749.
- 21Bazzi F, Rodriguez-Callejas JDD, Fonta C, et al. Brain segmentation from super-resolved magnetic resonance images. 2019 Fifth International Conference on Advances in Biomedical Engineering (ICABME). 2019.
- 22Bazzi F, Mescam M, Basarab A, Kouamé D. On single-image super-resolution in 3D brain magnetic resonance imaging. In: 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). 2019; p. 2840-2843.
- 23Risser L, Sadoun A, Mescam M, et al. In vivo localization of cortical areas using a 3D computerized atlas of the marmoset brain. Brain Structure and Function. 2019; 224: 1957-1969.
- 24Pohl KM, Fisher J, Grimson WEL, Kikinis R, Wells WM. A Bayesian model for joint segmentation and registration. Neuroimage. 2006; 31: 228-239.
- 25Palazzi X, Bordier N. The marmoset brain in stereotaxic coordinates. In: The Marmoset Brain in Stereotaxic Coordinates. New York, NY: Springer; 2008. p. 1-59. https://doi.org/10.1007/978-0-387-78385-7_1.
10.1007/978-0-387-78385-7_1 Google Scholar
- 26Woodward A, Hashikawa T, Maeda M, et al. The Brain/MINDS 3D digital marmoset brain atlas. Scientific Data. 2018; 5: 180009.
- 27Liu C, Ye FQ, Yen C-C, et al. A digital 3D atlas of the marmoset brain based on multi-modal MRI. Neuroimage. 2018; 169: 106-116.
- 28Iriki A, Okano HJ, Sasaki E, Okano H. The 3-Dimensional Atlas of the Marmoset Brain: Reconstructible in Stereotaxic Coordinates. Japan: Springer; 2018.
10.1007/978-4-431-56612-0 Google Scholar
- 29Tan C, Guan Y, Feng Z, et al. Deepbrainseg: automated brain region segmentation for micro-optical images with a convolutional neural network. Front Neurosci. 2020; 14: 179-191.
- 30De Feo R, Shatillo A, Sierra A, Valverde JM, Gröhn O, Giove F, Tohka J. Automated joint skull-stripping and segmentation with Multi-Task U-Net in large mouse brain MRI databases. NeuroImage. 2021; 229: 117734. https://doi.org/10.1016/j.neuroimage.2021.117734.
- 31Zhao N, Basarab A, Kouamé D, Tourneret JY. Joint segmentation and deconvolution of ultrasound images using a hierarchical Bayesian model based on generalized Gaussian priors. IEEE Trans Image Process. 2016; 25: 3736-3750.
- 32Bahrami K, Rekik I, Shi F, Shen D. Joint reconstruction and segmentation of 7T-like MR images from 3T MRI based on cascaded convolutional neural networks. International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. 2017; p. 764-772.
- 33Toma A. Joint Super-Resolution/Segmentation Approaches for the Tomographic Images Analysis of the Bone Micro-Architecture. PhD thesis, Universit1é de Lyon; 2016.
- 34Li Y. Joint Super-Resolution/Segmentation Approaches for the Tomographic Images Analysis of the Bone Micro-Structure. PhD thesis, Universit1é de Lyon; 2018.
- 35Fessler JA. Model-based image reconstruction for MRI. IEEE Signal Process Mag. 2010; 27: 81-89.
- 36Ayasso H, Mohammad-Djafari A. Joint NDT image restoration and segmentation using Gauss-Markov-Potts prior models and variational Bayesian computation. IEEE Trans Image Process. 2010; 19: 2265-2277.
- 37Mansouri M, Mohammad-Djafari A. Joint super-resolution and segmentation from a set of low resolution images using a Bayesian approach with a Gauss-Markov-Potts prior. Int J Signal Imaging Syst Eng. 2010; 3: 211-221.
10.1504/IJSISE.2010.038017 Google Scholar
- 38Hastings WK. Monte Carlo sampling methods using Markov chains and their applications. Biometrika. 1970; 57: 97-109.
- 39Neal RM. MCMC using Hamiltonian dynamics. In: S Brooks, A Gelman, G Jones, XL Meng, eds. Handbook of Markov chain Monte Carlo, vol. 2. United Kingdom: Chapman and Hall/CRC Handbooks of Modern Statistical Methods CRC Press; 2011, p. 2.
10.1201/b10905-6 Google Scholar
- 40Pereyra M, Dobigeon N, Batatia H, Tourneret JY. Estimating the granularity coefficient of a Potts-Markov random field within a Markov chain Monte Carlo algorithm. IEEE Trans Image Process. 2013; 22: 2385-2397.
- 41Carmi E, Liu S, Alon N, Fiat A, Fiat D. Resolution enhancement in MRI. Magn Reson Imaging. 2006; 29: 304.
- 42Greenspan H, Oz G, Kiryati N, Peled S. MRI inter-slice reconstruction using super-resolution. Magn Reson Imaging. 2002; 20: 437-446.
- 43Corbineau MC, Kouamé D, Chouzenoux E, Tourneret JY, Pesquet JC. Preconditioned P-ULA for joint deconvolution-segmentation of ultrasound images. IEEE Signal Process Lett. 2019; 26: 1456-1460.



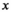 is modeled by a mixture of GGDs, the hidden label field
is modeled by a mixture of GGDs, the hidden label field 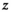 follows a Potts MRF and the mean
follows a Potts MRF and the mean 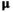 follows a Gaussian distribution. The parameters appearing in the boxes are fixed in advance
follows a Gaussian distribution. The parameters appearing in the boxes are fixed in advance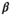 on the labels map produced by the JMCMC algorithm and that would be further used as prior maps for SPM segmentation
on the labels map produced by the JMCMC algorithm and that would be further used as prior maps for SPM segmentation