Sensitivity encoding for fast 1H MR spectroscopic imaging water reference acquisition
The copyright line for this article was changed on 29 September 2014 after original online publication.
Abstract
Purpose: Accurate and fast 1H MR spectroscopic imaging (MRSI) water reference scans are important for absolute quantification of metabolites. However, the additional acquisition time required often precludes the water reference quantitation method for MRSI studies. Sensitivity encoding (SENSE) is a successful MR technique developed to reduce scan time. This study quantitatively assesses the accuracy of SENSE for water reference MRSI data acquisition, compared with the more commonly used reduced resolution technique. Methods: 2D MRSI water reference data were collected from a phantom and three volunteers at 3 Tesla for full acquisition (306 s); 2× reduced resolution (64 s) and SENSE R = 3 (56 s) scans. Water amplitudes were extracted using MRS quantitation software (TARQUIN). Intensity maps and Bland-Altman statistics were generated to assess the accuracy of the fast-MRSI techniques. Results: The average mean and standard deviation of differences from the full acquisition were 2.1 ± 3.2% for SENSE and 10.3 ± 10.7% for the reduced resolution technique, demonstrating that SENSE acquisition is approximately three times more accurate than the reduced resolution technique. Conclusion: SENSE was shown to accurately reconstruct water reference data for the purposes of in vivo absolute metabolite quantification, offering significant improvement over the more commonly used reduced resolution technique. Magn Reson Med 73:2081–2086, 2015. © 2014 The Authors. Magnetic Resonance in Medicine Published by Wiley Periodicals, Inc. on behalf of International Society of Medicine in Resonance.
INTRODUCTION
1H Magnetic Resonance Spectroscopy (MRS) is a noninvasive technique which measures metabolite levels within a volume of interest 1-3. Several studies have demonstrated the value of this technique for investigating disorders of the central nervous system 4, with improvements in brain tumor 5, 6 diagnosis 6, prognosis 7, and characterization 5, 8 being particularly important due to the relatively poor outcome of this disease group.
The two most popular types of MRS investigation are single voxel spectroscopy (SVS) and MR spectroscopic imaging (MRSI). Single voxel spectroscopy (SVS) collects metabolic information from a single volume (voxel) of interest and is more commonly used than MRSI due to its shorter scan time 9 and relative ease of data collection and analysis. However, the restriction of information from a single location limits the number of appropriate clinical applications for the method. MR spectroscopic imaging (MRSI) or chemical shift imaging (CSI) is a multivoxel technique which can spatially map metabolite information throughout a predefined volume 10. This technique is practically promising for the investigation of diseases such as brain tumors, where tumor heterogeneity 11 and diffuse margins 12 are commonly observed features, with significant clinical interest. MRSI also offers advantages over SVS for investigating neurodegenerative diseases such as Alzheimer's 13 and neurometabolic disorders 4, 14; where the most clinically relevant brain area may not be known in advance.
Absolute quantification of metabolite levels is challenging but offers advantages over simple metabolite ratios. First, ratios can become unstable when the denominator metabolite is present at low levels; and, second, an overall reduction or increase in tissue metabolism would be difficult to detect using ratios, because all metabolites may be equally affected. Absolute quantitation is most commonly performed by referencing metabolite signal amplitudes to the signal obtained from water which acts as an internal standard 15-17. This method has been shown to be effective for SVS and can be routinely performed due to a minimal increase in scan time (<20 s). However, the combined collection of metabolite and water reference data for MRSI results in significantly longer acquisition times, as both data sets require phase encoding for spatial localization 18. The additional time required for standard MRSI phase encoding may preclude absolute quantitation for routine clinical use 19.
In recent years, fast-MRSI methods have been developed to reduce the number of phase encoding steps and, therefore, scan time. Sensitivity encoding (SENSE) is a parallel imaging technique which reduces the k-space sampling density by exploiting known spatial sensitivity profiles of multiple receiver coils; allowing more rapid spatial encoding 10, 18. The amount of k-space sampling density reduction is defined by a reduction factor R 20, for example R = 3 represent a three times reduction in the number of phase encoding steps. The fully sampled information is then algorithmically reconstructed from the undersampled data from each coil and the corresponding sensitivity and noise profiles 10, 18. SENSE is particularly popular for reducing MRI scan times and presents significant advantages over other fast techniques as it can easily be incorporated within any existing MRSI pulse sequence, avoiding pulse sequence related SNR losses 10.
A simpler method for reducing MRSI scan time is known as reduced k-space acquisition (reduced resolution). This method samples fewer points in k-space, resulting in a lower resolution scan. To ensure these data can be used as a quantitation reference for higher resolution (water suppressed) scans, the matrix size is increased by zero-filling the outer rim of k-space, effectively interpolating the missing data in the spatial domain 9. On some platforms, reduced resolution data may be collected primarily to assist postprocessing steps such as phase-correction and lineshape distortion removal; however, here we focus on its use for absolute metabolite quantitation.
Spatial resolution is theoretically preserved in the SENSE method, with an associated loss of SNR associated with higher reduction factors 20. Because SNR is extremely high for water reference data we hypothesise that SENSE will be a superior method for rapid water reference MRSI data acquisition when compared with the reduced k-space acquisition (the current default for the Philips MRSI protocol). In this study, we compare the accuracy of the reduced resolution and SENSE R = 3 techniques for obtaining a fast-MRSI water reference scan. To evaluate the accuracy of the two fast methods—water amplitudes were compared with equivalent fully sampled MRSI water reference acquisitions. The fast method that provided water amplitudes closest to the fully sampled method was regarded as the most accurate. Data were collected from three volunteers and a standard MRS phantom (“braino”) to validate the proposed methodology.
METHODS
MRSI Data Collection
Data were collected from three healthy volunteers (aged between 20 and 25 years) and a MRS “braino” phantom containing 10 mM creatine hydrate, 2 mM choline chloride, 5 mM dl-lactic acid, 1 mL/L Gd-DPTA (Magnevist), 12.5 mM l-glutamic acid, 7.5 mM myo-inositol, 12.5 mM N-acetyl-laspartic acid (NAA), 0.1% sodium azide, 56 mM sodium hydroxide (NaOH), and 50 mM potassium phosphate monobasic (KH2PO4) 21. This study had full ethical approval and informed consent.
All MR scanning was performed on a 3 Tesla (T) Philips Achieva TX MR system with a 32-channel head coil at Birmingham Children's Hospital, UK. An initial T1 weighted three-dimensional Fast Low Angle SHot (FLASH) MRI 1mm isotropic reference scan was obtained for MRSI grid positioning. All MRSI scans were manually positioned above the corpus callosum (see Figure 1a for example) with the following acquisition parameters: field of view (FOV) matrix size 15 × 13; voxel size 13 mm × 13 mm × 13 mm; TE = 35 ms; repetition time = 2 s; half-echo acquisition mode. In each case PRESS localization was used to excite a 6 × 6 voxel region (78 mm × 78 mm × 13 mm) centrally within the FOV. This corresponded to a fully exited 5 × 5 voxel region with a ½ voxel margin outside the PRESS excitation region. Because NMR visible water concentrations are fairly constant throughout the head (∼40 M) we intentionally included partially excited voxels (7 × 7 region) in the analysis to ensure the fast methods can be used to accurately reconstruct nonuniform water distributions. In all MRSI examinations, the points of k-space outside an elliptical boundary were not sampled to reduce acquisition time 22. A k-space hamming filter was applied as a postprocessing step to reduce ringing artifacts. No additional averaging was performed during k-space acquisition.
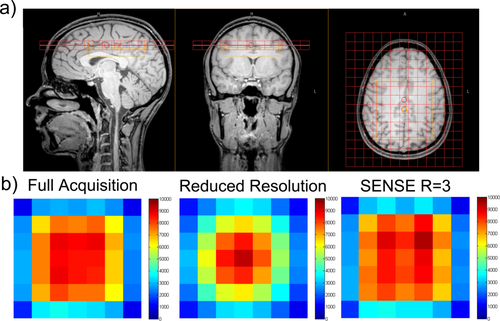
a: MRSI geometry for volunteer 2 including 6 × 6 voxel PRESS volume which is collocated with the shim box shown in orange. b: Water amplitude maps extracted from volunteer 2 for full acquisition; reduced resolution acquisition; and SENSE acquisition (R = 3).
Initial studies comparing SENSE with a full MRSI acquisition highlighted a systematic difference in the scaling factor between SENSE and no SENSE acquisitions. This was due to a rescaling step required for SENSE reconstruction. To allow a true comparison between the SENSE R = 3 and reduced resolution method, SENSE reconstruction was also enabled for the reduced resolution and full acquisition scans with a speedup factor of R = 1 (no undersampling). This resulted in a consistent scaling factor between the full acquisition, reduced resolution and SENSE methods. For the remainder of the manuscript the following terms will be used to describe the MRSI acquisitions used: “full acquisition (no-SENSE)” will refer to the fully sampled MRSI grid without SENSE scaling, 153 phase encoding steps = 5 min 6 s; “full acquisition” will refer to the fully sampled MRSI grid with sensitivity scaling (R = 1), 153 phase encoding steps = 5 min 6 s; “reduced resolution acquisition” will refer to a 2× reduction in k-space sampling for both dimensions with sensitivity scaling (R = 1), 32 phase encoding steps = 1 min 4 s; and “SENSE acquisition” will refer to the standard SENSE reconstruction with a 3× reduction k-space sampling for both dimensions (R = 3), 28 phase encoding steps = 56 s.
For the patient and phantom studies MRSI scans were performed in succession with identical field of view and PRESS geometries within a session. The following scans were collected for both patient and phantom data: full acquisition no-SENSE; full acquisition (R = 1); SENSE R = 3 acquisition; and reduced resolution acquisition.
Data Analysis
MRSI spectra were exported from the scanner workstation into the DICOM format and imported into the TARQUIN MRS quantitation software 23 for analysis. Water amplitudes were extracted from the time-domain data by back extrapolating the initial part of the FID to time = 0 (details of method given in Wilson et al) 23. Water amplitudes for each voxel within the FOV were then imported into MATLAB R2012a for statistical analysis. Grid maps of intensity were generated for no SENSE, SENSE R = 1, SENSE R = 3 and reduced resolution ×2. All voxels within the PRESS box (VOI) and the surrounding partially exited region (7 × 7 grid) were used for subsequent analysis. Bland Altman plots and associated statistics 24 were used to measure the agreement between the different MRSI acquisition protocols.
RESULTS
Scaling of Sensitivity Encoded MRSI Data in Comparison with the Full Acquisition (no-SENSE)
Before the comparison between the faster MRSI methods, a quantitative analysis between the full acquisition and full acquisition (no-SENSE) data were performed to validate the subsequent use of full acquisition (R = 1) as a valid comparator data set. Figure 2 shows a Bland Altman plot between the full acquisition methods for a phantom and volunteer data set. The mean difference and standard deviations in amplitude between the no-SENSE and SENSE R = 1 full acquisitions were −0.13 ± 3.62% for a phantom and −0.48 ± 5.51% for volunteer data (Fig. 2).
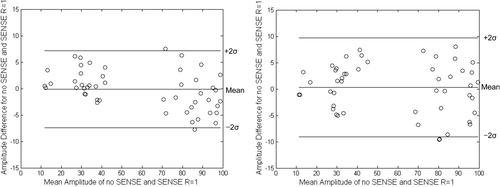
Water amplitude Bland-Altman plots to assess variance between full acquisition (SENSE R = 1) and full acquisition no-SENSE for phantom (a) and volunteer (b) data.
Comparison between Full Acquisition and Fast-MRSI Techniques
A 6 × 6 voxel VOI was excited using PRESS (Fig. 1a) and water amplitude (Fig. 1b) maps were produced for all MRSI acquisition methods for both phantom and volunteer data. A visual inspection of the water amplitude intensity maps (Fig. 1b) show that SENSE R = 3 gives a more accurate reconstruction of the full acquisition data when compared with the reduced resolution technique.
Table 1 shows the mean differences and standard deviations between the full acquisition versus SENSE and reduced resolution acquisitions for the phantom and all the volunteers. These were used to quantitatively determine any systematic or randomly distributed differences between full acquisition and the fast-MRSI acquisitions. For SENSE acquisition a mean difference in water amplitude of 2.31 ± 2.09% was observed for a phantom, low variability is observed about the mean. The reduced resolution technique showed greater variability with a mean difference and standard deviation in water amplitude for the phantom of 11.1 ± 11.1%.
| Water amplitude/ max amplitude (%) | |||
|---|---|---|---|
| Mean difference | SD | ||
| SENSE R=3 Vs full acquisition | Braino phantom | 2.31 | 2.09 |
| Volunteer 1 | 0.75 | 2.95 | |
| Volunteer 2 | 2.49 | 3.40 | |
| Volunteer 3 | 3.17 | 3.21 | |
| Reduced resolution x2 Vs full acquisition | Braino phantom | 11.11 | 11.09 |
| Volunteer 1 | 9.87 | 10.16 | |
| Volunteer 2 | 11.19 | 11.12 | |
| Volunteer 3 | 9.95 | 10.96 | |
In general the results are consistent between the volunteers and similar errors are seen between the phantom and volunteer results for the reduced resolution and SENSE acquisitions. The average mean and standard deviation was 2.1 ± 3.2% for the SENSE acquisition and 10.3 ± 10.7% for the reduced resolution technique. Therefore, we can conclude that the SENSE acquisition is approximately three times more accurate than the reduced resolution technique, both in terms of systematic bias and randomly distributed differences. These statistics are consistent with the visual differences seen in the intensity map in Figure 1.
Water Amplitude MRSI Reproducibility
Full acquisition (R = 1) and SENSE (R = 3) MRSI scans were acquired in duplicate for both the “braino” phantom and a volunteer data set to measure the reproducibility of the scans. The mean difference and its standard deviation between repeats were determined.
For the full acquisition (R=1) data a mean difference of −0.02 ± 0.26% was found for water amplitude for the “braino” phantom, and −0.45 ± 0.8% for volunteer data. An increase of SENSE factor to R=3 produced a mean amplitude difference between “braino” scans of 0.26 ± 0.34% and a mean difference of −0.94 ± 1.1% for the volunteer. In both cases, the reproducibility was better for the phantom, suggesting that subject motion causes an additional random error of less than 1%.
Phantom Metabolite Concentrations Using SENSE and Reduced Resolution Water Reference Data
SENSE (R = 3) and reduced resolution water reference data were used to estimate absolute metabolite concentrations found in the braino phantom. The same fully sampled water suppressed data were used for both analyses to ensure any differences could be attributed to the water reference data. The following metabolites were present in the phantom at known concentrations: 2.5 mM total N-acetyl-laspartic acid (tNAA), 10 mM total Creatine (tCr), 2 mM total Choline (tCho), and 12.5 mM glutamate (Glu). Mean metabolite concentrations were extracted from the central 5 × 5 voxel region, and estimated values were found to be more accurate for the SENSE water data in comparison with the reduced resolution technique. For SENSE : tNAA concentration = 11.98 ± 1.14 mM, tCr = 9.75 ± 1.49 mM, tCho = 2.72 ± 0.47 mM, and Glu = 13.7 ± 1.07 mM. For the reduced resolution technique : tNAA concentration = 16.01 ± 2.71 mM, tCr = 12.92 ± 2.29 mM, tCho = 3.61 ± 0.72 mM, and Glu = 18.39 ± 3.42 mM. A consistent overestimation in concentrations was found with the reduced resolution technique (Fig. 3a) due to an incorrect reduction of the water amplitude at the PRESS box edges (Fig. 1). SENSE provides a more uniform metabolite distribution as expected with a phantom.
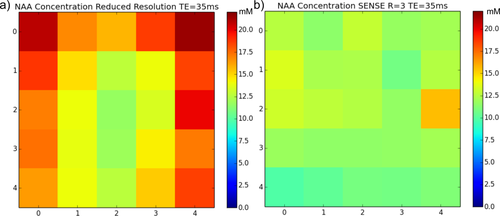
NAA concentration map for a phantom calculated from reduced resolution water reference data (a) and SENSE (R = 3) water reference data (b) for the central 5 × 5 voxel region.
Parietal White Matter Metabolite Concentrations Using SENSE and Reduced Resolution Water Reference Data
Absolute metabolite quantitation using the TARQUIN algorithm was performed on the volunteer data to demonstrate the feasibility of combining fully sampled water suppressed data with rapidly collected MRSI water reference data. Left and right parietal white mater voxels were analyzed for each of the three volunteers resulting in six voxels. Supporting Figure S1, which is available online, shows a typical example spectrum for this brain region. Table 2 shows the average volunteer metabolite concentrations for tNAA, tCr, tCho, and Glu calculated from the six voxels. Metabolite concentrations were determined using SENSE (R = 3) and reduced resolution water reference data. Concentrations calculated using the SENSE water data were found to be more consistent with those found in literature for healthy volunteers 25, 26 whereas the reduced resolution technique was found to over-estimate these values.
| Parietal white matter SENSE R=3 | Parietal white matter reduced resolution | |||
|---|---|---|---|---|
| Metabolite | Mean concentration (mM) | SD (mM) | Mean concentration (mM) | SD (mM) |
| tNAA | 8.53 | 0.30 | 10.62 | 0.34 |
| tCr | 5.56 | 0.16 | 6.89 | 0.15 |
| tCho | 1.14 | 0.14 | 1.41 | 0.19 |
| Glu | 5.45 | 0.21 | 6.77 | 0.18 |
DISCUSSION
The purpose of this study was to validate the use of SENSE for collecting fast-MRSI water reference data of the brain for the absolute quantification of metabolites. SENSE and reduced resolution methods were compared with: (i) equivalent fully sampled data sets from volunteer and phantom data; (ii) known metabolite concentrations from phantom data, and (iii) metabolite concentrations from healthy volunteers. In each of these cases it was found that SENSE MRSI offers a significant improvement in accuracy over the reduced resolution method. To the best of the author's knowledge this is the first study to have validated the use of SENSE for this purpose.
The improvement in accuracy of SENSE over the reduced resolution method is expected, because reduced resolution involves zero-filling in k-space, which is equivalent to interpolation. For a given spatial dimension the point spread function of the reduced resolution method is inversely proportional to the number of the acquired phase encoding points (for a fixed field of view)—rather than the number of zero-filled points. The SENSE method is based on k-space reconstruction rather than interpolation, and, therefore, provides improved resolution over a time-equivalent reduced resolution method.
Figure 1 shows that the SENSE method outperforms the reduced resolution method, particularly at the edges of the PRESS excitation region. Whilst this study has not directly tested the accuracy of the methods on greatly heterogeneous tissue water concentrations distributions, the heterogeneity caused by the PRESS excitation boundary is a valid model for testing accuracy. We expect that a similar investigation into heterogeneous tissue water distributions, for example in pathology, would yield comparable results. Furthermore, accurate quantitation (and, therefore, water amplitude measures) close to the boundary of the PRESS excitation region are desirable, and it is clear from this work that SENSE outperforms the reduced resolution method in these regions.
Absolute metabolite quantitation is generally preferred over using metabolite ratios because, in the case of ratios, the source of variation for a given ratio cannot be determined as to whether it is due to a relative increase in one metabolite or a decrease in the other 15. Whilst fast and accurate absolute metabolite quantitation for MRSI was the main goal for this work; water reference data can also be used to determine the “proton resonance frequency shift” for the purposes of noninvasive thermometry 27. Therefore, we anticipate this type of acquisition may also be useful for providing absolute temperate maps across the brain.
Pattern recognition performed directly on spectral data has been used previously as an alternative to absolute quantitation. These methods offer the advantage of being straightforward to implement because statistical methods, such as independent component analysis, are readily available. However, these widely available methods are not currently optimized for MRS specific issues such as variable line widths, unstable baselines, and residual water. Therefore, pattern recognition applied to the results from absolute quantitation offers the best of both approaches and has been demonstrated in pediatric and adult brain tumor studies 28, 29.
As with all MR methods, the protocol used in this study represents a compromise between scan-time, spatial resolution (voxel size) and unwanted T1/T2 weighting. Whilst MRSI resolution is comparatively poor, the additional spectral dimension allows a unique noninvasive view on tissue metabolism that makes it well suited for the investigation of certain diseases, in particular cancer. The MRSI parameters chosen for this study represent typical values for clinical MRSI where short scan times are particularly important. Whilst partial volume effects and incomplete relaxation are inevitable, clinically useful information can still be obtained from voxel sizes and repetition times used in this study.
The data quality of metabolite information acquired using SENSE has previously been assessed by several groups concluding that no significant losses were found in comparison with full acquisition data 18, 30, 31. However, Van Cauter et al found high SNR losses in lower concentration metabolites such as myo-inositol when using a SENSE factor of R = 3.6 19. Therefore, to preserve these lower concentration metabolites we propose the use of SENSE R = 1 for collection of metabolite data and SENSE R=3 for water reference data collection reducing the scan time for both metabolite and water reference data from 10 min 16 s to 6 min 6 s.
In addition to SENSE, two other methods have been shown to provide a promising acceleration of MRSI data acquisition: (i) EPI based methods such as PEPSI 32, 33 and (ii) compressed sensing 34. In this work, we chose to focus on SENSE due to its wider commercial availability and, therefore, greater clinical relevance. However, the underlying strategy of sacrificing SNR (rather than resolution) for reducing scan time is generic. Unlike MRSI water suppressed scans for metabolite signal measurement, MRSI water reference data have an extremely high SNR; therefore, it is likely that other fast methods that sacrifice SNR for a reduction in scan time will be similarly successful. In particular, compressed sensing, in isolation or combination with SENSE, may offer further reductions in scan time and would, therefore, make an interesting extension to this work.
CONCLUSIONS
SENSE has been shown to be approximately three times more accurate than the reduced resolution approach for acquiring fast MRSI water reference maps. Differences in water amplitude levels using SENSE were found to be less than 4% when compared with an equivalent full resolution acquisition. These findings validate the use of SENSE MRSI to obtain accurate water reference data in a feasible time frame for the purposes of absolute metabolite quantitation in a clinical setting.
ACKNOWLEDGMENTS
This work was supported by the Engineering and Physical Sciences Research Council [EP/F50053X/1] and National Institute for Health Research.