TSSK3, a novel target for male contraception, is required for spermiogenesis
Saman Nayyab and María G. Gervasi contributed equally to this study.
Abstract
We have previously shown that members of the family of testis-specific serine/threonine kinases (TSSKs) are post-meiotically expressed in testicular germ cells and in mature sperm in mammals. The restricted post-meiotic expression of TSSKs as well as the importance of phosphorylation in signaling processes strongly suggest that TSSKs have an important role in germ cell differentiation and/or sperm function. This prediction has been supported by the reported sterile phenotype of the TSSK6 knock-out (KO) mice and of the double TSSK1/TSSK2 KO. The aim of this study was to develop KO mouse models of TSSK3 and to validate this kinase as a target for the development of a male contraceptive. We used CRISPR/Cas9 technology to generate the TSSK3 KO allele on B6D2F1 background mice. Male heterozygous pups were used to establish three independent TSSK3 KO lines. After natural mating of TSSK3 KO males, females that presented a plug (indicative of mating) were monitored for the following 24 days and no pregnancies or pups were found. Sperm numbers were drastically reduced in all three KO lines and, remarkably, round spermatids were detected in the cauda epididymis of KO mice. From the small population of sperm recovered, severe morphology defects were detected. Our results indicate an essential role of TSSK3 in spermiogenesis and support this kinase as a suitable candidate for the development of novel nonhormonal male contraceptives.
1 INTRODUCTION
Male reproduction depends on the production of morphologically differentiated spermatozoa in the testis in a process known as spermatogenesis. Spermatogenesis is organized into three phases: (1) proliferative or spermatogonial phase in which spermatogonia undergo mitotic divisions and generate a pool of spermatocytes; (2) meiotic phase generating haploid spermatids; and (3) post-meiotic phase known as spermiogenesis (Sharpe, 1994). In mammals, spermiogenesis is characterized by dramatic morphological changes that occur in the haploid spermatid. These changes include formation of the acrosome, condensation, and reorganization of the chromatin, elongation and species-specific reshaping of the cell, and the assembly of the flagellum (Cheng & Mruk, 2011; O'Donnell, 2014).
Given the importance of phosphorylation in cell signaling and differentiation, it is not surprising that protein kinases are involved in spermatogenesis (Sassone-Corsi, 1997). However, only a few of them are exclusively expressed in the testis (Jinno et al., 1993; Nayak et al., 1998; Shalom & Don, 1999; Toshima et al., 1998, 1999; Tseng et al., 1998; Walden & Cowan, 1993). Several kinases have testis-specific splicing variants; two of the most relevant cases are the protein kinase A catalytic subunit and casein kinase II alpha. Null mice of the testis-specific variants of these enzymes are sterile (Nolan et al., 2004; X. Xu et al., 1999). Other examples of testis-enriched or specific kinases are the t-complex kinase Smok (Herrmann et al., 1999), the recently described HIPK4 (Crapster et al., 2020), and members of the testis-specific serine kinase family (TSSK). The TSSK family has six members, TSSK1, 2, 3, 4, 5, and 6 (TSSK5 is a pseudo-gene in humans), which are all expressed post-meiotically during spermiogenesis (Bielke et al., 1994; Hao et al., 2004; Kueng et al., 1997; Li et al., 2011; Visconti et al., 2001; Zuercher et al., 2000) (for a recent review see Salicioni et al., 2020). The members of the TSSK family have high homologies in their kinase domains, with TSSK1 and TSSK2 presenting the highest homology, followed closely by TSSK3, TSSK4, and TSSK6 (Hao et al., 2004; Li et al., 2011). The predicted importance of TSSKs has been confirmed using knock-out (KO) mice models. Male, but not female, Tssk6 (aka SSTK) KO mice (Spiridonov et al., 2005) and Tssk1/Tssk2 double KO (Shang et al., 2010) are sterile without exhibiting somatic abnormalities. In addition, Tssk4 KO mice are sub-fertile (Wang et al., 2015).
On the other hand, TSSK3 is a less studied member of this kinase family. This kinase was independently reported by Zuercher et al. (2000) and by our group (Visconti et al., 2001). It is also exclusively expressed post-meiotically in the testis (Li et al., 2011; Visconti et al., 2001) and remains in mature sperm (proteomic studies in Hwang et al., 2019). In the present work, we used CRISPR/Cas9 to produce TSSK3 loss-of-function mice. Tssk3−/− males F0 generation were sterile after several trials. Therefore, Tssk3-/- homozygous females were mated with wild-type C57BL/6J mice to produce three independent Tssk3−/− mouse transgenic lines. After multiple rounds of breeding, our results indicate that, each of these homozygous mouse models present a male (but not female) sterility phenotype. In all three Tssk3−/− lines, the number of mature sperm was very low. The few mature sperm have morphological defects and were completely immotile. Interestingly, round cells positive for the acrosomal marker peanut agglutinin (PNA) were present in the cauda epididymis. This sperm phenotype is significantly more dramatic than the one obtained from Tssk6−/−, or the combined Tssk1/2−/−. Altogether, our results indicate that TSSK3 is indispensable for male reproduction and validate the use of this kinase as a male contraceptive target.
2 RESULTS
2.1 TSSK3 is conserved among vertebrates
Although the TSSK family as well as their testis-restricted expression have been conserved through evolution, TSSK3 has evolved by duplication only in vertebrates. We analyzed the level of conservation of the TSSK3 protein in vertebrates at both the amino acid and gene structure levels. The representative alignment of 15 vertebrate TSSK3 (Figure 1) shows the high levels of conservation in this protein. The sequence GVAHRDLKCEN containing the putative active site, with an Asp residue in position 134 (mouse) is conserved among vertebrates, denoting the presence of a strong functional constraint in the evolution of this domain. Analogously, the sequence IGEGTYSKV (positions 16–24 in mouse), which is inferred to be the ATP-binding site, is conserved among vertebrates. In mammals, 225 of 268-9 residues are identical (84%), while 17 of 43 substitutions (40%) occurred between residues with similar physicochemical properties.
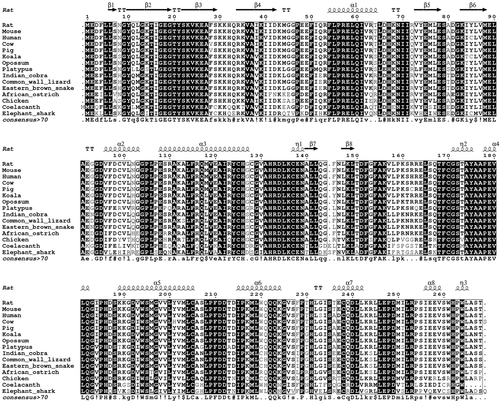
The average pairwise p-distance within placentals was 0.032 (SD 0.005), and between placentals and coelacanth and cartilaginous fish were 0.309 (0.028) and 0.325 (0.028), respectively. From a total of 28 sequences analyzed, 21 (75%) produce a protein of 268 residues, while 93% (all but two sequences) fall within the range 266–270 (Table S1). The exceptions are the ferret (Mustela putorius) and the kakapo (Strigops habroptila), which are 225 and 289 residues long, respectively. In the ferret, the N-terminus is shortened due to the loss of the first 20 nucleotides of the coding sequence, which is contained in a single exon and depicts high levels of conservation compared with other placentals TSSK3. In contrast, the sequence of the kakapo has a longer and highly divergent N-terminus (69 residues) after which it is very similar to any other bird TSSK3. With the exception of the ferret, all vertebrates TSSK3 CDS are encoded by two exons. The ferret and the coelacanth have a third exon, which corresponds to the 5′-untranslated region. All evidence indicates that TSSK3 evolved under purifying selection, intensified in functional domains. The limited variation was acquired along internal branches leading to major vertebrate groups, denoting that even these adaptations occurred under a strong negative selection background.
We then performed an analysis to study whether natural selection affected TSSK3 evolution. The selection analysis revealed that TSSK3 evolved under a strong purifying selection pressure (Figure S1a-b). Site-based methods showed a predominant proportion of negatively selected codons over positively selected ones (Table S2A). Positive selection was significantly detected in three codons, in positions 31, 49, and 110 of the alignment. The first (codon 31) led to a Lys→His substitution in chicken, while the second (codon 49) went through three amino acid replacements: Lys→Pro in coelacanth, Lys→Glu in amniotes, and Glu→Asp in the african ostrich. Five aminoacid replacements occurred in the last positively selected codon detected (110) Gln→Glu in coelacanth, Gln→Arg in the ancestor of amniotes, a parallel replacement Arg→Ser in the ancestor of therian mammals (marsupials and placentals), and in squamates (snakes and lizards), while two additional replacements occurred in chicken (Arg → His) and platypus (Arg → Pro). Lineage-based methods revealed evidence of episodic diversifying selection in the branch leading to mammals, while in the mammalian clade was significant for selection intensification (Table S2B). No evidence of gene-wide episodic diversifying selection was detected in the TSSK3 phylogeny (Table S2B). These results indicate that TSSK3 evolved under strong purifying selection denoting its importance among vertebrates. It is worth noting that the Ensembl gene tree (Figure S1a) lacks amphibian species, which would denote a gene loss event in this group. Indeed, when performing a TBLASTN search against Tropical clawed frog Xenopus_tropicalis_v9.1 (Genomic sequence) using the TSSK3 amino acid sequence as query, the two highest hits are the genes pskh1 and pskh2, which encode for two nonhomologous protein serine kinases, also present in other vertebrates (including mammals). These results confirm that TSSK3 was lost in the branch leading to amphibians and has probably been functionally replaced by a paralog.
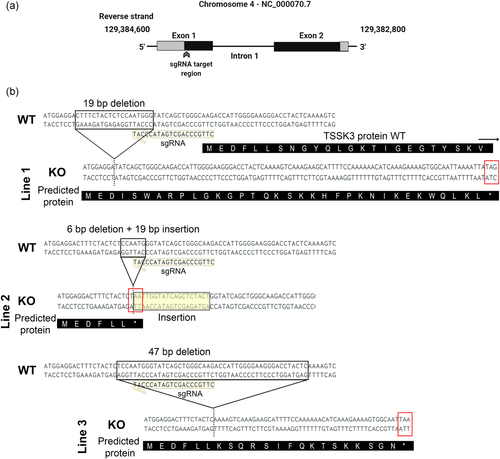
2.2 Generation ofTssk3KO mice using CRISPR/Cas9
The gene coding for Tssk3 maps to chromosome 4 in the mouse (Visconti et al., 2001) and has two exons. To evaluate the consequences of the absence of the Tssk3 gene, we used a CRISPR/Cas9 approach with a sgRNA targeting the Tssk3 gene ORF in exon 1 (Figure 2a). Upon microinjection of the sgRNA and Cas9 mRNA into B6D2F1 zygotes, embryos were in vitro cultured, and blastocysts were transferred to pseudo-pregnant CD1 female mice resulting in delivery of 10 pups, five males and five females. Males homozygous from the F0 generation were sterile (not shown). Therefore, to generate Tssk3−/− mice colonies, three females homozygous F0 mice were mated with wild type (WT) C57BL/6J mice, and three independent KO mouse lines were generated. Each of them resulted in frameshift mutations and generated early stop codons (Figure 2b). The absence of the targeted region in each case was confirmed by both polymerase chain reaction (PCR) and Sanger sequencing.
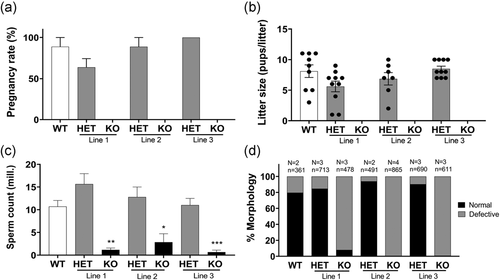
2.3 Tssk3−/− males but not females present a sterility phenotype
To evaluate the reproductive phenotype, Tssk3+/+, Tssk3+/−, and Tssk3−/− males from the three independent KO lines were mated with C57BL/6J WT females, alternatingly over a period of 2 months, as explained in the Methods section. All of them produced an equivalent number of plugs indicating no defects in reproductive behavior. While WT and heterozygous mice achieved similar pregnancy rates and litter size, homozygous Tssk3−/− males were sterile (Figure 3a,b). On the other hand, females Tssk3−/− were fertile with pregnancy rates and litter size comparable to WT mice (data not shown). Phenotypically, no differences were observed on epididymis size between Tssk3+/+, Tssk3+/−, and Tssk3−/− mice. However, the number of sperm obtained from the cauda epididymis was greatly reduced in the Tssk3 KO animals (Figure 3c). Almost all those sperm cells showed morphology abnormalities, such as malformed heads, when compared to heterozygous and WT (Figures 3d and 4a) and they were completely immotile. We noticed the presence of a considerable number of round cells in cauda sperm suspensions from the Tssk3 KO lines. Interestingly, a high percentage of the observed round cells was stained with PNA (Figure 4a) suggesting that haploid round spermatids in different stages of differentiation had reached the cauda epididymis. The severe morphological abnormalities presented by the Tssk3 KO sperm were studied in more detail by transmission electron microscopy (TEM) (Figure 4b). We found that ~30% of the KO sperm lacked some axonemal components and associated structures. Malformed heads with a detached acrosome were also detected (Figure 4b).
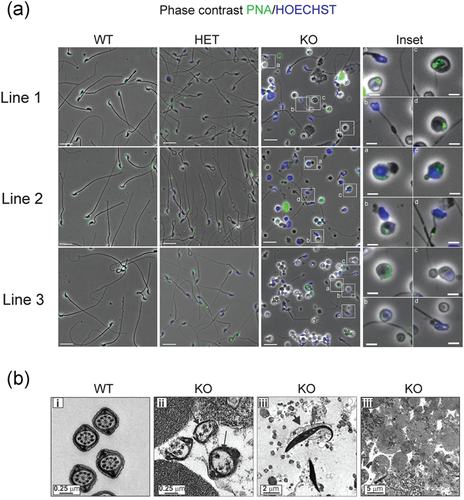
2.4 Spermiogenesis is affected in Tssk3−/− mice
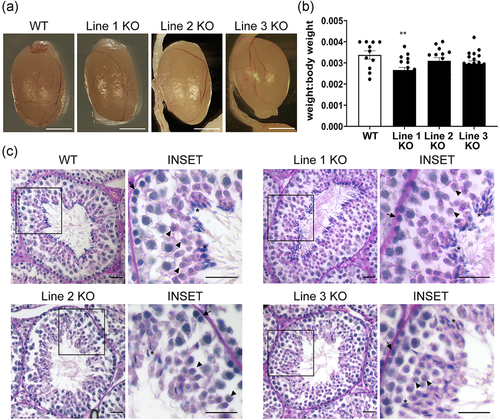
Results from the comparative phenotypic characterization studies of WT and Tssk3 KO mice revealed spermatogenic defects in the KO animals without obvious problems in testicular appearance (Figure 5a) or weight (Figure 5b). To gain further insight, histological evaluation of the WT and KO testis sections was performed using PAS-hematoxylin staining of paraffin-embedded tissues (Figures 5c and S2). Evaluation of about 400 or more seminiferous tubules in WT and KO testis sections from each line showed a normal distribution pattern for all the stages of spermatogenesis, and morphology of the germ cells at most of the spermatogenesis stages was indistinguishable between WT and KO testis sections. Additionally, diameters of seminiferous tubules appeared comparable between WT and KO testis sections. However, a further assessment of the maturing spermatids showed a distinct abnormality in step 15/16 spermatids in KO seminiferous tubules. In contrast to WT, step 15 and 16 spermatids in KO testis sections either failed to line up in a circle at the lumen (Lines 2 and 3) or displayed an abnormal organization pattern (Line 1) (Figure 5c). These results suggest that spermatogenesis is blocked at the stage VII/VIII in TSSK3-null mice and indicated that TSSK3 plays a crucial role during spermiogenesis.
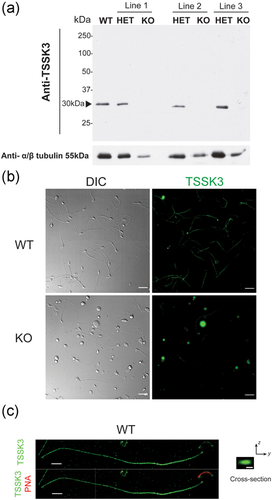
2.5 TSSK3 localizes to the sperm tail in mature sperm
Finally, having the KO mouse model presented an excellent opportunity to investigate TSSK3 localization. A polyclonal anti-TSSK3 antibody (AP7246a) recognized a band of 30 KDa in sperm from Tssk3+/+ and Tssk3+/− mice but not in sperm from Tssk3−/− mice (Figure 6a). When used in immunofluorescence experiments, this antibody stained the sperm flagellum of WT but not the one from Tssk3−/− sperm (Figure 6b). It is worth noticing that suspensions from Tssk3−/− mice cauda epididymis depicted some staining in non-sperm structures and in a small fraction of heads and flagella (Figure 6b). Whether this signal results from either nonspecific staining or is a truncated form of TSSK3 remains to be clarified. A more detailed localization of TSSK3 in WT sperm by three-dimensional structured illumination microscopy (3D-SIM) indicated that TSSK3 localized to the center of the sperm flagellum suggesting an axoneme localization (Figure 6c) which is consistent with the observed alterations in axoneme structures detected by EM.
3 DISCUSSION
All members of the TSSK family are almost exclusively expressed post-meiotically in testicular germ cells. Among family members, Tssk1 was cloned in 1994 (Bielke et al., 1994) by reverse-transcription PCR (RT-PCR) using degenerated primers targeting regions conserved in protein kinases catalytic domains (Hanks & Quinn, 1991). Tssk2 was cloned subsequently by the same group using the sequence of Tssk1 in low stringency hybridization screening (Kueng et al., 1997). Independently of each other, the same researchers and our group cloned Tssk3 (aka Stk22D) using a similar degenerate primer approach designed to discover novel testis-specific kinases (Visconti et al., 2001; Zuercher et al., 2000). The highly abundant expression of TSSKs in the testis and the finding that they were developmentally regulated gave rise to the hypothesis that these kinases play a role in spermiogenesis or later on in mature sperm function. A second study by our group (Hao et al., 2004), in which the human homologues of the mouse Tssks were cloned, demonstrated that human TSSKs also exhibit testis-specific expression when analyzed using quantitative PCR, Northern and dot blots of over 50 human tissues. More recent papers published on TSSKs include the finding that Tssk6 null (Spiridonov et al., 2005) and the double Tssk1/Tssk2 null (Shang et al., 2010; B. Xu et al., 2008) KO mutant mice are sterile. Interestingly, each TSSK isoform presents differential localization in sperm. TSSK1 and TSSK4 are present in the anterior head as well as in the flagellum. TSSK2 and TSSK6 are found only in the actin-rich post-acrosomal region (Li et al., 2011; Sosnik et al., 2009). In addition, our group and others have developed in vitro assays that have increased the understanding of the TSSKs' mode of regulation (Bucko-Justyna et al., 2005; Hawkinson et al., 2017; Jaleel et al., 2005; Li et al., 2011).
The high levels of conservation observed in amino acid sequences as well as in gene structure (e.g., conservation of the number of exons and introns) among vertebrates, a group which arose about 525 mya (million years ago) (Table S1), are indicative of a strong selective pressure acting on this gene. The selection analyses performed in this study further confirmed that TSSK3 evolved under strong purifying selection, denoting its crucial role in fitness maintenance. Only 3 (1%) out of 271 analyzed codons showed evidence of positive selection, while up to 195 (72%) codons showed significant evidence of purifying selection (Table S2B). Codons 49 and 110 (48 and 109 in mouse, respectively) represent shared hotspots of positive selection across long evolutionary times, as supported by the occurrence of multiple amino acid replacements along the vertebrates tree. Branch-site methods revealed the occurrence of episodic diversifying selection (EDS) in the branch leading to mammals, a fact that is congruent with the evolution of several lineage-specific reproductive traits in this group. Nonetheless, after the occurrence of mammalian-specific novelties, TSSK3 has undergone a process of selection intensification, which is indicative of the presence of strong functional constraints. Taken together, our results confirm that since its origin, TSSK3 has been involved in crucial functions such as reproduction or survival, producing severe phenotypes in mutants.
In the present work, we used the CRISPR/Cas9 system to generate three independent mouse colonies lacking Tssk3. As shown in Figure 2, two of the mutations were caused by deletion of 19 and 47 base pairs, respectively. The third one was caused by an deletion of 6 base pairs combined with an insertion of 19 base pairs; in all cases, the reading frame was disrupted and generated early stop codons. Although we cannot completely discard the possibility that a downstream methionine was used, even if this occurred, the resultant protein would lack the first two regions of the kinase catalytic domains, rendering the enzyme inactive. In the three colonies, male Tssk3−/− mice were unable to impregnate WT females upon mating. On the other hand, Tssk3−/− females were fertile and gave similar number of pups as their WT siblings.
Although in our previous manuscript we failed to detect TSSK3 protein in sperm (Li et al., 2011), a recent proteomic analysis identified TSSK3 in sperm extracts (Hwang et al., 2019). Here, we repeated those western blots using harsher cell lysis conditions and were able to detect a protein of the right molecular weight that disappeared in sperm from Tssk3−/− mice. One possibility to explain these results is that, in the previous work, we used milder, nonreducing conditions to prepare sperm extracts. Here, we used reducing agents as a first step during the cell lysis (the sperm DNA is decondensed, forming a viscous solution which is very difficult to pipette) followed by sonication to minimize any protein-containing clumps; this may also have increased the possibility of pulling out TSSK3 that might be bound to cytoskeletal structures, before analysis. In addition, the validated antibody localized TSSK3 in the sperm flagellum; importantly, sperm from Tssk3 KO mice served as negative controls.
As mentioned, male but not female Tssk3−/− mice are sterile. This phenotype is not surprising; the number of mature sperm collected from the epididymis obtained by swim-out was negligible. When instead of using the sperm swim-out method, the cauda epididymis was squeezed, different cell types were recovered. Using this method, we were able to obtain mature sperm; however, the number of differentiated cells was very low in all cases. In addition to differentiated sperm, many round cells were observed. Many of these cells were positive when stained with PNA, suggesting that these round cells are spermatids under different stages of differentiation. Contrary to either the double Tssk1 KO/Tssk2 KO, the Tssk6 KO, or the Tssk4 KO mice models, the number of sperm in the Tssk3 KO is severely reduced and the few sperm cells obtained are completely immotile. Moreover, the presence of round cells in the cauda epididymis provides a strong evidence for aberrant spermatogenesis in Tssk3 null mice. This finding is consistent with the observed spermatogenic arrest in KO testis as evidenced by the failure of step 15/16 spermatids to align, forming a circle, in the lumen of stage VII/VIII tubules. Overall, these observations support the hypothesis that TSSK3 plays a critical role during late spermiogenesis and/or release of sperm into the lumen. Further work is warranted to determine the precise biological function of TSSK3; we expect that the Tssk3−/− mouse lines will provide valuable tools to gain deeper insights into the process of spermiogenesis and male fertility.
Importantly, TSSK family members have been proposed as male contraceptive targets. Among the properties that make this kinase family attractive for this purpose, the following should be mentioned: (1) The male sterility phenotypes of the Tssk6−/− (Spiridonov et al., 2005), the double Tssk1−/−/Tssk2−/− (Shang et al., 2010) and the Tssk3−/− (this manuscript) mice, added to the male subfertile phenotype of Tssk4 KO (Wang et al., 2015), indicate that TSSKs play essential roles in male reproduction without affecting other tissues; (2) TSSKs are developmentally regulated during spermatogenesis and are only expressed post-meiotically in germ cells (Li et al., 2011; Salicioni et al., 2020), suggesting that therapeutic reversibility would be possible since spermatogonia stem cells would not be affected; (3) sequences of TSSK family members are evolutionary conserved in vertebrates and invertebrates (Boutet et al., 2008; Marcello et al., 2012; Salicioni et al., 2020), consistent with a conserved role in male reproduction; (4) The importance and “druggability” of kinases in many diverse physiological processes suggests that specific inhibitors of TSSKs may be found, and would be expected to block spermatogenesis and/or fertilization; (5) because TSSKs remain in mature sperm after spermiogenesis (Li et al., 2011), it is possible that they have a role in sperm function. Therefore, specific TSSK inhibitors could be also useful in females to prevent fertilization.
All TSSK members have the potential to be used as contraceptive targets. However, only TSSK6 has been individually validated using loss-of-function KO technology. Although the double Tssk1/Tssk2 KO mouse model is sterile, this mouse model alone is not sufficient to validate these kinases as contraceptive targets because it is not clear the extent by which each TSSK (1 and/or 2) contributes to the sterile phenotype. On the other hand, TSSK4 and TSSK5 are not considered human contraceptive targets; Tssk4 KO males are not completely sterile and TSSK5 is a pseudogene in humans. Evidence presented in this study provides strong indication for TSSK3 as a new validated male contraceptive target.
4 METHODS
4.1 Chemicals
Reagents were obtained from various sources and details are provided below, under the subsections pertaining to each experimental procedure. Bovine serum albumin (A-7906, for blocking nonspecific binding), Tween-20 (P-7949), Immobilon-P PVDF membrane (Millipore), and poly-l-lysine (P-8920) were purchased from Sigma-Aldrich. Paraformaldehyde (16% solution, EM Grade), and β-mercaptoethanol were purchased from Fisher Scientific. Triton X-100 and 30% acrylamide/Bis solution were obtained from BioRad. Peroxidase-conjugated anti-mouse IgG (Jackson Immunoresearch) and peroxidase-conjugated anti-rabbit IgG (GE Healthcare) were used. AlexaFluor488-conjugated PNA lectin, AlexaFluor488-conjugated anti-rabbit IgG, Hoechst 333452, and gentamicin (10 mg/ml) were purchased from Invitrogen (Thermo Fisher Scientific). Anti-TSSK3 rabbit polyclonal antibodies AP7246a and AP17810b were obtained from Abcepta. A rabbit polyclonal antibody against alpha/beta-tubulin was obtained from Cell Signaling Technology.
4.2 Evolutionary protein sequence analysis
The gene structure of the Tssk3 orthologs in vertebrates was analyzed. A total of 28 vertebrate TSSK3 protein sequences (Supporting Information File S1) were aligned with Clustal Omega (Madeira et al., 2019). Uncorrected p-distances were calculated with MEGA X (Kumar et al., 2018). Additionally, an alignment with 15 representative species was obtained, generating a graphical representation with ESPript 3 (Robert & Gouet, 2014). Sequence similarities were calculated considering residue physicochemical properties (%Equivalent option; similarity threshold = 0.7). Secondary structure was inferred from the related 2r0i.1.A Serine/threonine-protein kinase MARK2 Par-1 mutant (Rat) crystal structure available in the SWISS-MODEL Repository (Bienert et al., 2017).
To assess if natural selection affected the evolution of TSSK3, we employed codon-based and lineage-based Bayesian and Maximum Likelihood approaches to estimate rates of non-synonymous (dN) to synonymous substitutions (dS). A 27 vertebrate TSSK3 protein alignment (the highly divergent kakapo sequence was excluded) (Supporting Information File S2) was used as reference for converting nucleotide alignments into codon alignments employing PAL2NAL (Suyama et al., 2006). The codon alignment and a species phylogenetic tree were uploaded to the Datamonkey webserver (Weaver et al., 2018) and selection was inferred using the following codon-based methods. Single sites under selection were identified using Single Likelihood Ancestral Counting (SLAC) (Kosakovsky Pond & Frost, 2005), Fixed Effects Likelihood (FEL) (Kosakovsky Pond & Frost, 2005), adaptive Branch-Site Random Effects Likelihood (aBSREL) (Smith et al., 2015), Mixed Effects Model of Evolution (MEME) (Murrell et al., 2012), as well as Fast Unconstrained Bayesian AppRoximation (FUBAR) (Murrell et al., 2013). SLAC infers dN and dS at each codon position comparing observed and expected rates based on a single ancestral sequence reconstruction. FEL estimates and compares dN and dS independently on a per-site basis. aBSREL performs also a per-site dN and dS estimation but allows for overall dN/dS (ω) heterogeneity. MEME aims to detect single sites evolving under positive selection along particular branches. FUBAR enables larger numbers of site classes and identifies positively selected sites using a Bayesian framework. Significance thresholds for selection tests were p ≤ 0.10 for SLAC, FEL, and MEME, p ≤ 0.05 for aBSREL, and posterior probability ≥0.90 for FUBAR. Selection at each domain was also tested using tree-based methods at the Datamonkey server: RELAX and BUSTED. Given two subsets of branches in a phylogeny, RELAX (Wertheim et al., 2015) determines whether selective strength was relaxed or intensified in one of these subsets relative to the other. BUSTED (Branch-Site Unrestricted Statistical Test for Episodic Diversification) (Murrell et al., 2015) tests for evidence of EDS in at least one site and one branch of the phylogeny.
4.3 Generation of Tssk3 knock-out mouse line by CRISPR/Cas9
A Tssk3 KO allele was generated in the Animal Models Core Facility at the Institute for Applied Life Sciences (IALS) (University of Massachusetts-Amherst), according to the animal protocol #2018-0005 approved by the Institutional Animal Care and Use Committee. Briefly, B6D2F1 (C57BL/6J × DBA2J) female mice (8–10 weeks old) were superovulated with 7.5 IU pregnant mare serum gonadotropin (PMSG; BioVendor), followed by 7.5 IU human chorionic gonadotropin (hCG; Sigma-Aldrich) 48 h later. Superovulated females were mated with B6D2F1 males and euthanized at 20 h post-hCG injection for zygote collection from the oviducts. Oviductal ampullae were dissected to release zygotes, and cumulus cells were removed by pipetting in M2 medium containing hyaluronidase (EMD Millipore). Zygotes were then washed and microinjected in M2 medium (EMD Millipore). A volume of 5-10 pL with single guide RNA (2 ng/µl, targeting Exon 1 of Tssk3 on CTTGCCCAGCTGATACCCAT) and Cas9 mRNA (3.3 ng/µl; TriLink BioTechnologies, L-7206) was microinjected into the cytoplasm of zygotes. Microinjected zygotes were cultured in KSOM medium (EMD Millipore) at 37°C in a humidified atmosphere of 5% CO2/5% O2 balanced with N2. After 3 days of in vitro culture, early blastocysts (E3.5) were transferred into the recipients (E2.5) by using Nonsurgical Embryo Transfer (ParaTechs Corporation, #60010). Delivered pups were genotyped by using the primers with Forward: 5′-AAGAAGGAGGCGCTACACAG-3′ and Reverse: 5′-GGCCCTCCCATCTTGTCTAT-3′. All PCR amplicons from all delivered pups were also TA cloned (Thermo Fisher Scientific, K204040) for Sanger sequencing (Psomagen Inc.) for the selection of the founders.
4.4 In vivo fertility test
Male mice, with either wild-type, Tssk3+/− or Tssk3−/− genotype, were mated with wild-type females (≥2-month-old) (1 male:2 female ratio) in individual cages, and checked for vaginal plugs every morning. Once plugs were identified, female mice were separated from the male, transferred to a new cage, and monitored for pregnancy and pups. Males that successfully plugged a female were given a day of rest before being placed back in a cage with two other female mice. If the plugged female gave birth, pups were then counted; after 3 weeks, the mother was recovered and used for another mating activity. To obtain biological replicates, at least three male mice of each genotype were used, and three or more vaginal plugs were allowed per male over approximately a 2-month period of intermittent mating cycles. Percent pregnancy was calculated as the number of litters per number of plugs per male; the average percent of pregnancy of different males for each genotype was then obtained.
4.5 Animals usage and spermatozoa collection
Animals handling and euthanasia were in accordance with the Animal Care and Use Committee (IACUC) guidelines of UMass-Amherst (protocol #2019-0008). Cauda epididymal mouse sperm samples were obtained from both epididymides by squeezing using dissecting tweezers and collected into 1 ml of modified Toyoda–Yokoyama–Hosi (mTYH) medium. This medium contains the following compounds (concentrations are given in parenthesis): NaCl (119.3 mM), KCl (4.7 mM), CaCl2 × 2H2O (1.71 mM), KH2PO4 (1.2 mM), MgSO4 × 7H2O (1.2 mM), glucose (5.56 mM), Na pyruvate (0.51 mM), HEPES (20 mM), and gentamicin (10 mg/ml). This medium does not support mouse sperm capacitation. After squeezing, epididymides tissue debris was discarded, and sperm cells were counted using a hemocytometer. Wildtype and Tssk3+/− sperm cells were diluted 1:100 before counting.
4.6 Testes to body weight
Tssk3+/+, Tssk3+/−, or Tssk3−/− genotyped animals were sacrificed by cervical dislocation and immediately put on a scale to obtain the body mass. After the whole animal was weighed, each testis was dissected and weighed. Relative testis over body weight was calculated as the average of left and right testes over the bodyweight of each animal and then averaged for each genotype.
4.7 PAS-hematoxylin staining of testes
Whole testis tissues were collected for fixation, tissue dehydration, and paraffin embedding from adult WT-, Line 1 HET- and KO-, Line 2 HET- and KO-, Line 3 HET- and KO-male mice as previously described (Tourzani et al., 2018). Tissue samples were sectioned at 7.5-µm-thick slices, lifted onto Superfrost Plus positively charged glass slides (Fisherbrand) and left to dry at 37°C overnight before being stored at −20°C. Paraffin removal was performed by two consecutive incubations with 100% xylene for 15 min at room temperature. Slides were then rehydrated by incubation first in decreasing ethanol concentrations (100%, 95%, and 70%) for 5 min at room temperature, followed by a wash with filtered phosphate-buffered saline (PBS). PAS-hematoxylin staining was performed by incubation with 1% periodic acid (Fisherbrand) for 30 min at room temperature, then washed for 10 min in running water. Next, slides were incubated for 40 min in Schiff's reagent (Fisherbrand) at room temperature, then washed for 10 min in running water. Slides were finally counterstained with Mayer's hematoxylin (Sigma) for 3 min at room temperature, then washed for 15 min in running water. Slides were dehydrated in increasing ethanol concentrations (70%, 95% and 100%) for 5 min and cleared in xylene. Slides were then mounted with Cytoseal-60 (Richard-Allen Scientific). Brightfield images were taken using a ×40 objective (Nikon, LWD Phase, NA 0.55) in an inverted microscope (Nikon Eclipse T300) equipped with a RGB illumination system and an Andor Zyla camera.
4.8 Sperm morphology analysis
Sperm cells were obtained by squeezing, pelleted by centrifugation at 150g for 5 min, and incubated in 1 ml of 4% paraformaldehyde at room temperature for 10 min on a rocker. After removing the fixative by a second centrifugation at 150g for 5 min, cells were washed with 1 ml of PBS and resuspended in PBS (0.5–1 ml) depending on cell concentration. Then, 30 µl of the cell suspensions were added to poly-l-lysine treated #1.5 Corning coverslips (Fisher 12-553-464) and let air-dry. Samples were permeabilized with 0.5% v/v Triton X-100 in PBS for 5 min and washed with 0.1% Tween 20 in PBS (T-PBS) three times for 5 min each. Cells on coverslips were then treated with 1:100 Hoechst 33342 and 1:100 AlexaFluor488-conjugated PNA lectin in T-PBS (1 h at room temperature in the dark). After an hour incubation, coverslips were washed three times with T-PBS 5 min each in the dark. Coverslips were mounted on Global Diamond White Glass microscope slides with Vectashield (Vector Labs H-1400). The coverslip-slide was then sealed with nail polish along the edges. Phase-contrast and fluorescence images were taken with a ×40 LWD phase objective (NA 0.55; Nikon) with an inverted microscope (Nikon Eclipse TE300).
4.9 TSSK3 protein extraction and western blot analysis
After squeezing, sperm samples were collected by centrifugation and washed with 1 ml PBS. For extraction of TSSK3, cells were first resuspended in 0.5% sodium dodecyl sulphate (SDS) (in PBS). After the addition of β-mercapthoethanol (5% final concentration), protein samples were boiled for 10 min, and then sonicated for 10 sections (three times each) on ice. After centrifugation at 14,000 rpm, supernatants (and corresponding insoluble fractions) were saved at −20°C until further analysis. Protein concentration in cell lysates was determined using the BCA protein assay kit (by Pierce, from ThermoFisher), and protein extracts (30–50 µg of protein per lane) were separated by SDS–polyacrylamide gel electrophoresis (12% gels) and electro-transferred to PVDF membranes. After treatment with 5% fat-free milk in TBS-Tween20 (Tris-HCl 20 mM, NaCl 150 mM, pH 7.5 containing 0.1% Tween 20), to block nonspecific binding, immunoblotting was conducted with polyclonal anti-TSSK3 AP7246a at a concentration of 2ug/mL in 1% fat-free milk, followed by HRP-labeled antirabbit IgG in 1% fat-free milk/TBS-Tween20 at a concentration of 1:10,000. Detection of TSSK3 signal was enhanced with ECL prime. Equal loading was checked by blot-stripping and reprobing with an antibody against alpha/beta-tubulin.
4.10 Immunolocalization of TSSK3 in mouse sperm
Cauda epididymal sperm from wild-type, Tssk3+/− or Tssk3−/− genotype mice were retrieved by tissue squeezing in 1 ml of mTYH medium. Cells were centrifuged at 150 g for 5 min, resuspended in 1 ml 4% paraformaldehyde, and incubated at room temperature for 10 min on a rocker. After 10 min, the cell suspensions were centrifuged at 150g for 5 min and washed with 1 ml of PBS. Cells were then resuspended in 0.5–1 ml of PBS depending on cell concentration. Thirty microliters of cells were added to poly-l-lysine #1.5 Corning coverslips at room temperature until air dry. Cells were permeabilized by the addition of 0.5% v/v Triton X-100 in PBS for 5 min and washed three times with 0.1% Tween 20 in PBS (T-PBS) 5 min each. Cells were blocked by 10% BSA in T-PBS for 1 h at room temperature and then treated with 10 µg/ml of anti-TSSK3 AP7246a antibody in 1% BSA overnight at 4°C in the dark. The next day, cells were washed three times for 5 min each with T-PBS and treated with secondary anti-rabbit IgG AlexaFluor 488 (1:1000) and PNA (1:100) in T-PBS for 1 h in the dark at room temperature. Samples were washed five times with T-PBS for 5 min each and then mounted on Global Diamond White Glass microscope slides with Vectashield (H-1000). Using an inverted microscope (Eclipse TE300; Nikon) epifluorescence and DIC images were taken with a ×60 PlanApo/DIC objective (NA 1.49; Nikon).
4.11 3D-SIM
For TSSK3 labeling and SIM, sperm samples from WT animals were fixed and incubated with primary and secondary antibodies as described above for immunofluorescence. Three-dimensional dual-color SIM images were captured using a Nikon A1R-SIMe microscope with a Nikon PlanApo 100× NA1.49 objective. Image reconstruction for each channel was performed in NIS elements software (Nikon). Full projections of the reconstructed Z-stacks (0.2 µm) were created using the software Fiji (Schindelin et al., 2012).
4.12 TEM
Sperm samples were prepared for TEM following standard protocol adapted from Chemes (2013). Briefly, sperm were fixed with 2.5% Glutaraldehyde in 0.1 M Cacodylate Buffer, pH 7.4 for 15 min and washed twice in 0.1 M Cacodylate Buffer, pH 7.4, Specimens were then post-fixed by incubation with 1% osmium tetroxide, dehydrated by immersion in a graded series of alcohol solutions and embedded in Epon resin. Ultra-thin sections (90 nm) were cut and stained with uranyl acetate and lead citrate. Sections were analyzed with a JEOL USA JEM-1400 Flash TEM microscope at the VCU Massey Cancer Center Microscopy Core Facility.
4.13 Statistical analysis
Statistical analyses were performed using the Prism 8.4.3 software (by GraphPad). Data are expressed as mean ± SEM. All data were verified to accomplish the parametric assumptions of homogeneity of variances and normality. Variables were compared by one-way analysis of variance, and the statistical significance between groups was determined by Tukey's post-hoc test. Significant levels were considered when *p < 0.05, **p < 0.01, or ***p < 0.001.
ACKNOWLEDGMENTS
This study was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development 5P50HD093540 (to P.E.V. and G.I.G.); and by the Male Contraceptive Initiative Sokal Award MCI # 2020-304 (to P.E.V.) and a Trainee Fellowship MCI # 2020-B02 (to S.N). The authors would also like to thank NIH for partial support from National Research Service Award T32 GM108556 (Biotechnology Training Program funding to D.A.T.). The VCU Massey Cancer Center Microscopy Core Facility is partially supported by the NIH-NCI Cancer Center Support Grant P30 CA016059.
CONFLICT OF INTERESTS
Dr. Visconti and Dr. Salicioni own equity interest in Sperm Capacitation Technologies Inc., a company with goals in improving assisted reproductive technologies. The other authors have no conflict of interest to declare.