Identification and Peptidomic Profiling of Exosomes in Preterm Human Milk: Insights Into Necrotizing Enterocolitis Prevention
Abstract
Scope
Human breast milk has been shown to prevent necrotizing enterocolitis (NEC). Although exosomes have been identified in breast milk, their function and components have not been fully addressed. This study is conducted to elucidate the differences in peptidomic complexities between preterm and term milk exosomes.
Methods and results
Breast milk samples are collected from healthy lactating mothers who have delivered term and preterm infants. Exosomes are separated and quantified. The protective effects of purified exosomes against NEC are investigated both in vitro and in vivo. The peptidomic complexities in term and preterm milk exosomes are analyzed by iTRAQ LC-MS/MS to screen differentially expressed exosomal peptides. Preterm milk exosomes administration significantly enhances proliferation and migration of intestinal epithelial cells compared with term milk exosomes. A total of 70 peptides are found to be significantly modulated in preterm milk samples compared to term milk samples. Of these, 47 peptides are upregulated, and 23 peptides are downregulated. Bioinformatics analysis suggests several potential regulatory roles of the altered peptides in intestinal epithelial cell function.
Conclusion
These results reveal the differences for the first time in peptidomic complexities between preterm and term milk exosomes. Milk exosome administration might be a promising prevention for NEC.
1 Introduction
Breast milk not only provides ideal nutrition for the newborns, but it also has various bioactive components such as secretory immunoglobulins (particularly sIgA), immune competent cells (such as leukocytes), and antimicrobial factors (such as lysozymes, oligosaccharides, and antimicrobial peptides). It is considered to influence the development of the neonatal immune and gut systems.1 Breast milk feeding results in a reduced incidence of necrotizing enterocolitis (NEC), which is one of the leading causes of mortality among neonates, especially in premature infants.2 The benefits of breast milk feeding probably include developing the immune system, promoting intestinal immunity,3 killing luminal pathogens, and reducing the incidence of gastrointestinal food allergy.4 Although these effects of breast milk have been observed, the mechanism by which the components in milk protects neonates from NEC is still not yet clear.
Exosomes are small, endocytic vesicular bodies (30–200 nm) present in various body fluids and are especially highly present in breast milk.5, 6 Milk-derived exosomes encapsulate various types of cargos, such as proteins, mRNAs, and microRNAs (miRNAs), and protect them from enzymatic and non-enzymatic degradation.7 Therefore, milk exosomes bear important biological functions in mediating cell-to-cell communication among distant cells.8 Rat milk–derived exosomes have been found to promote intestinal epithelial cell proliferation, and stimulate intestinal stem cell activity, providing a possible explanation for why breast milk feeding is beneficial in preventing NEC.6 A recent study has found that bovine milk-derived exosomes could prevent the development of NEC by enhancing goblet cell activity.9 It is worth noting that milk from mothers who delivered preterm infants significantly differs from the milk of those delivering term infants in that it contains higher concentrations of carbohydrates, fat, and energy, in proportion to the decreasing gestational age of the infant.10 Although the exosomal miRNA profile has been explored,1 differences between the other contents of human preterm and term milk exosomes have received limited attention. Recently, the multifunctional properties of milk-derived bioactive peptides have been increasingly acknowledged. Numerous effects have been attributed to milk-derived biologically active peptides including antimicrobial, anti-inflammatory, anti-oxidative, antithrombotic, antihypertensive, and anti-obesity properties.11 It could have a positive impact on human metabolism and physiology, either directly or through enzymatic hydrolysis.12 For example, the KVLPVP peptide from casein effectively decreased blood pressure in hypertensive rats after oral administration.13 Hence, to better understand milk composition and lactation physiology regarding NEC prevention, it is important to unravel the milk exosome peptidomics of human preterm and term milk.
In this study, we aimed to analyze the effects of milk exosomes from mothers delivering preterm and term infants on human normal intestinal epithelial cells and NEC animal model, as well as to assess the differences in peptidomic complexities between preterm and term milk exosomes.
2 Experimental Section
2.1 Collection of Human Breast Milk
Breast milk samples were collected from healthy lactating mothers who had delivered term (≥37 weeks) and preterm infants (24-36 weeks) in Women's Hospital of Nanjing Medical University, Nanjing Maternity and Child Health Care Hospital. Colostrum samples were collected in the first 3 days after birth. All mothers were producing an excess of milk, which is normally discarded. These excess samples were collected for research purposes. Samples were stored at −80 °C until analysis. The collection procedures were approved by the Institutional Review Board at Women's Hospital of Nanjing Medical University, Nanjing Maternity and Child Health Care Hospital [Permission Number (2013)78].
2.2 Isolation and Purification of Human Milk Exosomes
Breast milk samples were collected in sterile 50 mL tubes and centrifuged twice at 3000 g for 15 min at 4 °C. The fat layer was removed and the supernatant was transferred to new tubes. Breast milk samples were then syringe-filtered with a 0.22 µm filter and prepared for ultracentrifugation at 42 000 rpm, 70 min at 4 °C. After centrifugation, supernatants were aspirated and the pellets were stored at −80 °C.
2.3 Cell Experiments
2.3.1 Cell Culture
Human normal intestinal epithelial FHC were purchased from American Type Culture Collection (Manassas, VA) and cultured in the recommended medium. This included DMEM: F12 medium (Gibco, Grand Island, NY, USA) and 10% exosome-free fetal bovine serum (Gibco, USA), supplemented with 100 U mL−1 penicillin and 100 µg mL−1 streptomycin (Gibco, USA); 25 mm HEPES (Thermo, Waltham, MA, USA); 10 ng mL−1 cholera toxin (Sigma-Aldrich, St Louis, MO, USA); 0.005 mg mL−1 insulin (Sigma-Aldrich, USA); 0.005 mg mL−1 transferrin; 100 ng mL−1 hydrocortisone (Sigma-Aldrich, USA) and 20 ng mL−1 human recombinant EGF (PeproTech, Suzhou, China). Cells were maintained in a 37 °C, 5% CO2 humidified incubator.
2.4 Immunofluorescence
Isolated milk exosomes (term and preterm) were labeled with PKH26 according to the manufacturer's protocol. The labeled exosomes were passed through 0.22 µm filter (Millipore, Billerica, MA, USA) and washed three times to remove excess dye. FHC cells were seeded and grown on 12-well plates for 24 h. Cells were incubated with serum-free medium for 2 h at 37 °C and then treated with PKH26 labeled exosomes (58 µg protein) for another 3 h and subsequently fixed and stained with DAPI. Cells were observed under a confocal microscope (Zeiss, Germany).
2.5 Scratch Closure Test
A total of 3 × 105 FHC cells were seeded into 6-well plates and scratched by a sterile pipette tip. Fresh serum-free culture medium containing Term-Exos or Preterm-Exos (154 µg of protein) were added. Scratched areas were observed using a microscope (Zeiss, Germany) at 0, 6, 12, and 24 h. The widths of the scratches were measured by the ZEN software (Zeiss, Germany).
2.6 Cell Proliferation Assay
The Cell Counting Kit-8 (CCK-8) (Dojindo, Tokyo, Japan) was used to determine cell proliferation according to the manufacturer's instruction. FHC cells were seeded on 96-well plates at a concentration of 1 × 103 cells per well and grown overnight. Then, the FHC cells were co-cultured with Term-Exos or Preterm-Exos (15.4 µg of protein) for 0, 12, and 24 h. The optical density (OD) value was measured at 450 nm using a microplate reader (BioTek Instruments Inc, Germany).
2.7 Animal Experiments
This study was approved by the Animal Research and Care Committee of Nanjing Medical University. Necrotizing enterocolitis (NEC) was induced in newborn rat pups as previously described with modification.14, 15 Newborn Sprague–Dawley rats were randomized into three groups. To induce NEC, 10-day-old rats were separated from their mothers and administered 50 µL g−1 body weight formula gavage [Similac Advance infant formula (Abbott Nutrition) : Esbilac canine milk replacer, 2:1] three times per day using a 24-French angiocatheter, as well as hypoxia (5%O2, 95%N2) for 5 min twice daily for 4 days. Milk exosome (200 µg mL−1) supplemented formulas were prepared and given to the indicated group. Control animals remained with their mothers and received breast milk. Gastrointestinal tract samples were harvested at day 4 and then fixed in 4% paraformaldehyde solution, embedded in paraffin, and 5-µm sections were stained with hematoxylin and eosin (HE) for microscopic evaluation. The BrdU immunostaining was performed as described.14
2.8 In Vivo Biodistribution of Milk Exosomes
The distribution of exosomes administered by oral gavage was investigated as previously described with modification.16-18 Briefly, filtered milk exosomes were labeled with fluorescent dye DiR (1 µm). Nude mice were administered a single dose of DiR-labeled exosomes (200 µg exo protein) by intragastric gavage (i.g.). The mice-administered free DiR served as control. The fluorescence intensities of whole mice were assessed using an in vivo imaging system (IVIS Spectrum, PerkinElmer, USA). Different organs were collected and imaged ex vivo as well.
2.9 Exosome Identification
The nanoparticle tracking analysis system (NanoSight LM10, Malvern Instruments Ltd., UK) and transmission electron microscopy (TEM) were used to determine particle size. Markers of exosomes were detected by western blotting using anti-human CD9 antibody (Abcam, Cambridge, MA, USA) and anti-human CD63 antibody (Abcam, USA).
2.10 Peptide Extraction
An equal volume of U2 (8 m Urea, 100 mm TEAB, pH 8.0) was added to an individual sample. The suspension was sonicated for 15 min on ice and then centrifuged at 4 °C, 13 000 g for 30 min. The supernatant was reduced with 10 mm DTT at 56 °C for 30 min and alkylated with 50 mm iodoacetamide (IAM) at room temperature for 30 min in the dark. The protein concentration was measured using the Bradford method. Then the sample solutions were filtered with 10 kDa MWCO filter tube by centrifugation (4 °C, 10 000 g) to remove proteins and peptides larger than 10 kDa. The flow-through was collected and the peptides were purified by C18 columns. The elutates were vacum dried and stored at −20 °C.
2.11 Sample Desalting and Labeling
After desalting, peptides were lyophilized and reconstituted in 20 µL of 0.5 m TEAB for peptide labeling. Peptides from samples were labeled with iTRAQ 8-plex kits (AB Sciex Inc., Framingham, MA, USA) according to the manufacturer's protocol. The labeled samples were combined and lyophilized. Next, iTRAQ-labeled samples were fractionated using a high-performance liquid chromatography (HPLC) system (Thermo DINOEX Ultimate 3000 BioRS) with a Durashell C18 (5 µm, 100 Å, 4.6 mm × 250 mm), and 12 fractions were collected.
2.12 LC-ESI-MS/MS Analysis Based on Triple TOF 5600 Plus
Each fraction was dissolved in 30 µL of 2% acetonitrile/0.1% formic acid and analyzed using TripleTOF 5600+ mass spectrometer coupled with the Eksigent nanoLC System (SCIEX, USA). Five microliters of the peptide sample was loaded onto a C18 trap column (5 µm, 100 µm × 20 mm), and eluted at 300 nL min−1 onto a C18 analytical column (3 µm, 75 µm × 150 mm) over a 90 min gradient. The two mobile phases were buffer A (2% acetonitrile/0.1% formic acid/98% H2O) and buffer B (98% acetonitrile/0.1% formic acid/2% H2O). For IDA (information-dependent acquisition), survey scans were acquired in 250 ms, and 30 product ion scans were collected in 100 ms per scan. MS1 spectra were collected in the range of 350–1500 m/z, and MS2 spectra were collected in the range of 100–1500 m/z. Precursor ions were excluded from reselection for 15 s.
2.13 Data Analysis
Protein identification and quantification were performed using ProteinPilot 4.5 Software (July 2012; AB Sciex). MS/MS spectra were searched against a UniProt -human protein database. The search parameters were set as follows: the instrument was a TripleTOF 5600, iTRAQ quantification, cysteine modified with iodoacetamide; biological modifications were selected as the ID focus, the Quantitate, Bias Correction, and Background Correction were checked for protein quantification and normalization. Detected peptide threshold [Unused ProtScore (Conf)]: 0.05 (10.0%); FDR Analysis tab checked. All identified proteins had an Unused Protscore of >1.3 (which corresponds to proteins identified with >95% confidence), as calculated by the software, and a global false discovery rate (FDR) of ≤1% determined at the protein level by the PSPEP algorithm. To be considered as being differentially expressed, proteins were required to have a p-value of ≤0.05, as calculated by the software.
2.14 Bioinformatics
General properties of the differentially expressed peptides were analyzed using ProtParam, the UniProt protein database, and peptigram. Functional classifications of the differentially expressed peptides were determined by GO enrichment analysis using the online DAVID tool. The protein–protein interaction networks were mapped using STRING. Wheel-like patterns of candidate peptides were visualized by helical wheel projections.
2.15 Statistical Analysis
Statistical analyses were performed with using GraphPad Prism 7 software. Student's t-test or one-way ANOVA was employed for statistical comparisons. The statistical significances were calculated as p-values, and p < 0.05 was considered statistically significant.
3 Results
3.1 Characterization of Human Breast Milk Exosomes
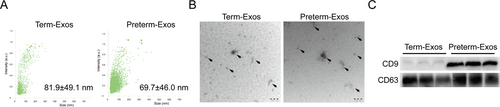
Both purified exosomes from term and preterm human breast milk had a mean size 50–100 nm in diameter as seen by nanosight tracking and electron microscopy analysis (Figure 1A,B), which is consistent with the previously known characteristics of exosomes. Exosome membranes are enriched in endosome-specific markers, such as CD9 and CD63. As shown in Figure 1C, CD9 and CD63 were both present in the purified exosomes.
3.2 Effects of Exosomes from Term and Preterm Human Breast Milk
More recently, breast milk has been shown to be protective against necrotizing enterocolitis (NEC).19 We hypothesized that this effect may be partially associated with exosomes. In order to exert effects on human normal intestinal epithelial cell line (FHC), we first investigated the exosome uptake in the cells. The co-incubation of PKH26-labeled exosomes with FHC cells led to readily internalized labeled exosomes, as indicated by the images taken at 2 h (Figure 2A). Compared with exosomes from term human breast milk (Term-Exos), the exosomes from preterm human breast milk (Preterm-Exos) significantly promoted the proliferation of FHC cells. The results of the in vitro scratch closure assay demonstrated that the migration of FHC cells increased at 6, 12, and 24 h in the presence of Preterm-Exos compared to Term-Exos (Figure 2C,D). To confirm the protective effects against NEC in vivo, milk exosomes were given to animals by oral gavage. As shown in Figure 2E, DiR-labeled exosomes could target intestines tissue and liver in vivo as early as 40 min post-administration. The NEC newborn rat models were induced by enteral administration of formula every 8 h, along with exposure to 5 min of hypoxia twice daily. On histological examination of the NEC rat model, injured areas occurring at the ileum showed a loss of villous integrity, which is consistent with the typical feature of NEC in preterm infants.20 Interestingly, administration of Preterm-Exos along with formula protected the villous integrity from injury in the NEC rat model (Figure 2F). Furthermore, the neonatal rats with NEC showed a significant reduction in enterocyte proliferation which was restored in Preterm-Exo-treated animals (Figure 2G).

3.3 Proteomic Analysis of Milk-Derived Exosomes by iTRAQ LC-MS/MS
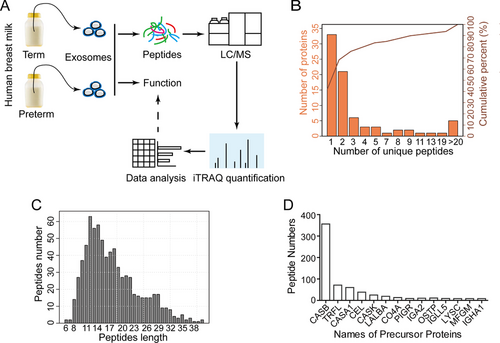
To understand such protective roles of Preterm-Exos, we performed a comparative peptidomic analysis of exosomes from term and preterm human breast milk (Figure 3A). A total of 719 peptides were identified in both the Term-Exo and Preterm-Exo groups. The number of proteins containing at least two unique peptides in this project was 46, accounting for 58.23% of the total proteins (Figure 3B). The average length of the identified peptides was 17.74, which was within a reasonable range of peptide length (Figure 3C). As shown in Figure 3D, the largest number of peptides is derived from the parent protein Casein beta (CASB).
3.4 Identification of Differentially Expressed Peptides in Preterm-Exos
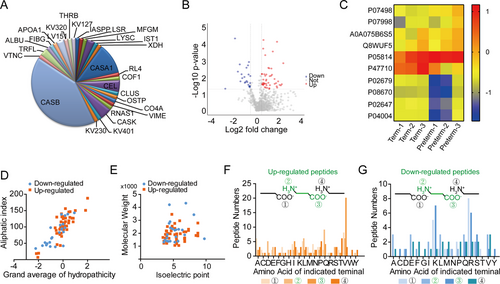
Peptides were identified to be differentially expressed when the iTRAQ ratios were >1.4-fold or <0.7-fold while p < 0.05 in the Preterm-Exo group. We found that 70 peptides originating from 28 parent proteins (Figure 4A) were differentially expressed in the Preterm-Exos group compared with that in the Term-Exos group, among which 47 peptides were upregulated, and 23 peptides were downregulated (Figure 4B). The detailed information of the most differentially expressed peptides is listed in Table 1. The typical expression profiles of the differential peptides are visualized as a heat map (Figure 4C). The distributions of the grand average of hydropathicity relative to the aliphatic index, and the isoelectric point (pI) relative to molecular weight (MW) were also investigated. The grand average of hydropathicity of most peptides ranged from −2 to 2, and the distribution of the aliphatic index mainly ranged from 50 to 150 (Figure 4D). The pI of most of the peptides ranged from 3.0 to 8.0, and the distribution of MW mainly ranged from 1 to 3 kDa (Figure 4E). Since the peptides usually derive from precursor proteins by enzymatic cleavage, we further explored the amino acids at the cleavage sites of differentially expressed peptides. In the upregulated group, the primary amino acids at the four cleavage sites were valine (V), glutamine (Q), tryptophan (W), and threonine (T) (Figure 4F), while in the downregulated group, the primary amino acids at the four cleavage sites were argnine (R), alanine/glutamic acid/leucine/glutamine/valine (A/E/L/Q/V), lysine (K), and valine (V) (Figure 4G).
| Peptide Sequences | Accessions | Gene | MW[KDa] | Fold change | P-value |
|---|---|---|---|---|---|
| Down-regulated peptides | |||||
| QIYPVTQPLAPVHNPIS | P05814 | CSN2 | 1874.17 | −6.8960 | 0.0476 |
| QTDEIKDTRNESTQNCVVAEPEKM | P47710 | CSN1S1 | 2766 | −3.7518 | 0.0000 |
| EKQTDEIKDTRNESTQNCVVAEPEK | P47710 | CSN1S1 | 2892.1 | −3.1175 | 0.0045 |
| ISNPTAHENYEKNNV | P47710 | CSN1S1 | 1729.82 | −2.7705 | 0.0016 |
| ENSHVQVPFQQ | P47710 | CSN1S1 | 1312.4 | −2.6573 | 0.0172 |
| LKKPVAFSDYIHPVCLPDRETAASLLQAGYK | P00734 | THRB | 3432 | −2.4770 | 0.0065 |
| FGSYCPTTCGIADFLSTYQTK | P02679 | FGG | 2303.59 | −2.3666 | 0.0111 |
| ISLPLPNFSSLNLR | P08670 | VIME | 1570.85 | −2.0701 | 0.0093 |
| SASEDIDFDDLSR | P53990 | IST1 | 1469.48 | −2.0131 | 0.0209 |
| Up-regulated peptides | |||||
| VNALALTLPFLAV | P07498 | CSN3 | 1341.66 | 24.6882 | 0.0130 |
| LHYVGFVPVIDGDFIPADPI | P19835 | CEL | 2184.52 | 5.5818 | 0.0014 |
| QKQPACHENDERPF | P07498 | CSN3 | 1698.83 | 5.0981 | 0.0254 |
| QSVLTQPPSVSAAPGQKV | P01701 | IGLV1-51 | 1794.04 | 4.7625 | 0.0299 |
| LDICSKNPCHNGGLCEEISQEVR | Q08431 | MFGE8 | 2544.85 | 4.4140 | 0.0279 |
| QVPFQQLNQLAAYPYAV | P47710 | CSN1S1 | 1950.22 | 4.0321 | 0.0173 |
| QSVLTQPPSVSAAPGQKVTI | P01701 | IGLV1-51 | 2008.3 | 3.7765 | 0.0410 |
| SQSGVALSPWVIQKNPL | P19835 | CEL | 1824.11 | 3.7431 | 0.0001 |
| DVVMTQSPLSLPV | P06310 | IGKV2-30 | 1385.64 | 3.6825 | 0.0340 |
| SALLQDNIADAV | P61626 | LYZ | 1229.35 | 3.5347 | 0.0219 |
| GFVPVIDGDFIPADPI | P19835 | CEL | 1671.91 | 3.2338 | 0.0243 |
| NTFVHEPLVDVQNV | P07998 | RNASE1 | 1610.79 | 3.2189 | 0.0124 |
| HQIYPVTQPLAPV | P05814 | CSN2 | 1462.71 | 2.8881 | 0.0003 |
| YPFVEPIPYGFLPQN | P05814 | CSN2 | 1781.04 | 2.7261 | 0.0069 |
| PTHQIYPVTQPLAPVHNPIS | P05814 | CSN2 | 2209.53 | 2.6354 | 0.0029 |
| PTHQIYPVTQPLAPVHNPISV | P05814 | CSN2 | 2308.67 | 2.5964 | 0.0079 |
| DGGFIYEAGLAPYKLRPV | P02788 | LTF | 1966.27 | 2.4050 | 0.0291 |
| EIVLTQSPGTLSLSPGERA | P01619 | IGKV3-20 | 1955.2 | 2.3799 | 0.0083 |
| LLLNPTHQIYPVTQ | P05814 | CSN2 | 1636.91 | 2.3181 | 0.0442 |
| VVLPVPQPEIMEVPKAKDTV | P05814 | CSN2 | 2189.64 | 2.2941 | 0.0028 |
| VLNQLCVLHEKTPV | P02768 | ALB | 1592.92 | 2.2940 | 0.0491 |
| EVQNQKQPACHENDERPFYQ | P07498 | CSN3 | 2460.62 | 2.1828 | 0.0027 |
| HQIYPVTQPLAPVHNPISV | P05814 | CSN2 | 2110.44 | 2.1809 | 0.0209 |
| IYPSFQPQPLIYPFVEPI | P05814 | CSN2 | 2148.53 | 2.0276 | 0.0420 |
3.5 Gene Ontology (GO) Enrichment and Association Network Analysis of Peptide Precursors
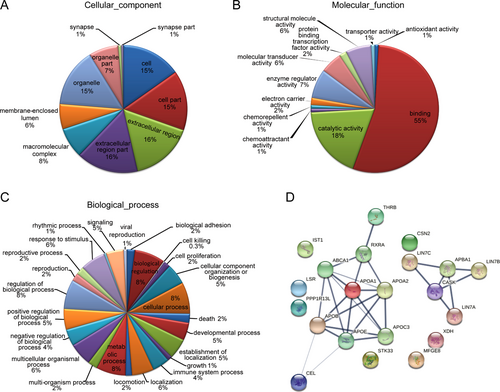
GO enrichment analysis of the identified peptide precursors in the Preterm-Exos group was applied in terms of their cellular compartments, molecular function, and biological process. We found that the cell, extracellular region, macromolecular complex, membrane-enclosed lumen, and organelle were the most highly enriched cellular components, confirming that the isolated exosomes were of an adequate purity (Figure 5A). Moreover, these peptide precursors were mainly involved in molecular functions such as binding, catalytic activity, enzyme regulator activity, structural molecule activity and molecular transducer activity (Figure 5B). For the biological process, these peptide precursors were major participants in the metabolic process, developmental process, immune system process, biological adhesion, and cell proliferation (Figure 5C). The association network analysis was used to visualize the interaction network of altered peptide precursors in the Preterm-Exo group (Figure 5D).
3.6 The Candidate Peptides with Potentially Bioactive Function
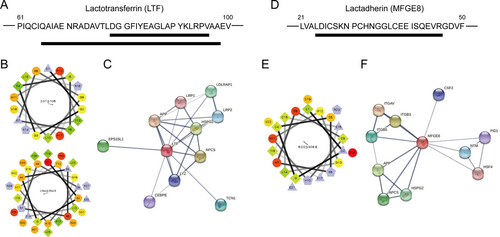
The differentially expressed peptides derived from the functional domains of their parent proteins are summarized in Table 2. We identified a total of 22 peptides located in the functional domains of their precursors, of which nine were downregulated and 13 were upregulated. When combined with previous studies, we found that the peptides derived from the Transferrin-like 1 domain of lactotransferrin (LTF) (Figure 6A) and the peptide derived from the EGF-like domain of lactadherin (MFGE8) (Figure 6D) might be potentially associated with the effects of milk exosomes on the FHC cells. In addition, helical wheel and association network analysis were used to illustrate the properties and potential function of LTF-derived peptides (Figure 6B,C) and MFGE8-derived peptide (Figure 6E,F).
| Peptide sequences | Protein | Position | Domain | Description | Modifications |
|---|---|---|---|---|---|
| Down-regulated peptides | |||||
| LKKPVAFSDYIHPVCLPDRETAASLLQAGYK | Prothrombin | 469–499 | 365–619 | Peptidase S1 | Disulfide bond |
| FGSYCPTTCGIADFLSTYQTK | Fibrinogen gamma chain | 41–61 | – | – | Disulfide bond |
| ISLPLPNFSSLNLR | Vimentin | 411–424 | 103–411 | IF rod | Phosphoserine |
| VSFLSALEEYTK | Apolipoprotein A-I | 251–262 | 68–267 | 10 X approximate tandem repeats | – |
| ASSIIDELFQDR | Clusterin | 183–194 | – | – | Interchain |
| ESKKEDLVFIFWAPESAPLK | Cofilin-1 | 93–112 | 4–153 | ADF-H | Phosphoserine/N6-acetyllysine/Phosphothreonine/Glycyl lysine isopeptide |
| DVWGIEGPIDAAFTR | Vitronectin | 198–212 | 158–250 | Hemopexin 1; Hemopexin 2 | – |
| ALLQDNIADAVAC | Lysozyme C | 101–113 | – | – | Disulfide bond |
| AIPVAQDLNAPSDWDSRGKDSYETSQLDDQSAETHSHK | Osteopontin | 204–241 | – | – | Phosphoserine/Phosphotyrosine |
| Up-regulated peptides | |||||
| QSVLTQPPSVSAAPGQKV | Immunoglobulin lambda variable 1–51 | 20–37 | 20–117 | Ig-like | Pyrrolidone carboxylic acid |
| LDICSKNPCHNGGLCEEISQEVR | Lactadherin | 24–46 | 24–67 | EGF-like | Disulfide bond/Phosphoserine |
| QSVLTQPPSVSAAPGQKVTI | Immunoglobulin lambda variable 1–51 | 20–39 | 20–117 | Ig-like | Pyrrolidone carboxylic acid |
| DVVMTQSPLSLPV | Immunoglobulin kappa variable 2–30 | 21–33 | 21–120 | Ig-like | – |
| SALLQDNIADAV | Lysozyme C | 100–111 | – | – | Disulfide bond |
| NTFVHEPLVDVQNV | Ribonuclease pancreatic | 72–85 | – | – | Disulfide bond |
| DGGFIYEAGLAPYKLRPV | Lactotransferrin | 79–96 | 25–352 | Transferrin-like 1 | – |
| EIVLTQSPGTLSLSPGERA | Immunoglobulin kappa variable 3–20 | 21–39 | 21–116 | Ig-like | – |
| VLNQLCVLHEKTPV | Serum albumin | 480–493 | 404–601 | Albumin 3 | Disulfide bond |
| DIQMTQSPSSLSASVGDRV | Immunoglobulin kappa variable 1–27 | 23–41 | 23–117 | Ig-like | – |
| QAIAENRADAVTLDGGFIYEAGLAPYKLRPVAA | Lactotransferrin | 66–98 | 25–352 | Transferrin-like 1 | – |
| APPAPIPPPAPSQSS | RelA-associated inhibitor | 583–597 | – | – | Phosphoserine |
| IQVTVSNPYHVV | Lipolysis-stimulated lipoprotein receptor | 90–101 | 86–234 | Ig-like V-type | – |
4 Discussion
Necrotizing enterocolitis (NEC) primarily affect premature neonates, leading to a high number of neonatal deaths per year.5 Many survivors of severe NEC are left with extremely high morbidity of short bowel syndrome and abnormal neurodevelopmental outcomes.21 Effective preventive and therapeutic strategies are desperately needed for this devastating condition. Evidence from epidemiological studies has shown that breast milk feeding for the preterm neonate is beneficial in reducing morbidity of NEC.22, 23 One of the leading potential mechanisms could be related to the rich source of exosomes in breast milk.24 Colin et al. reported that human breast milk-derived exosomes can protect intestinal epithelial cells (IECs) from oxidative stress induced cell death.5 Alison et al. found that rat milk-derived exosomes can promote intestinal epithelial cell proliferation and intestinal stem cell activity.6 In the present study, we isolated exosomes from the milk of mothers who delivered preterm and term infants. Nanoparticle tracking and western blot analysis demonstrated a greater concentration of exosomes in the preterm sample compared to that in the term sample. We found that both preterm and term milk exosomes can be taken up by intestinal epithelial FHC cells and located to the cytoplasm. Furthermore, the Preterm-Exo administration significantly enhanced FHC cell proliferation compared with the Term-Exo-treated group. Similarly, there was a significant increase in cell migration in the Preterm-Exo-treated group. Intestinal restitution is an important process for the maintenance of the epithelial barrier and mucosal integrity in response to injury. Restitution involves the migration of enterocytes from healthy areas to the injury sites, in which IECs proliferate to generate new enterocytes.25 It has been suggested that NEC is associated with a remarkable inhibition of enterocyte migration and proliferation, making the host uniquely susceptible to further injury, which finally leads to bacterial translocation.15 Thus, Preterm-Exo have a great potential for NEC prevention. Our in vivo study also demonstrated the protective effects of Preterm-Exo in a neonatal NEC animal model. The results of in vivo analysis revealed that milk exosomes administered by oral gavage are mainly taken up by the intestinal tract. Moreover, the rats that were administered the Preterm-Exos showed a reduction in the severity of NEC, suggesting a major role in maintaining the integrity of villous trophoblast and promoting enterocyte proliferation. In sum, our findings strongly suggest that Preterm-Exos may have potential protective effects against NEC by enhancing the proliferation and migration of intestinal epithelial cells.
To gain a better understanding of the protective role of Preterm-Exos, we performed a peptidomic analysis of Preterm-Exos, comparing them to milk exosomes fom mothers who delivered term infants. Pevious studies on cow's milk have revealed that the composition in milk exosomes, such as miRNAs, caters to infant's needs.1 Sarah et al. demonstrated that the exosomal miRNAs from both term and preterm milk are associated with inflammation, immune system regulation, cell growth, and development.26 Bioactive peptides derived from milk are initially found even in an inactive form within the sequence of the precursor molecules, but they can be released and influence numerous physiological responses.27 In addition, several bioactive peptides show two or more different biological activities and are multifunctional.28 To our knowledge, the differences in peptidomic complexities between preterm and term milk exosomes have not been studied, and the study of milk exosomal peptide profiling on NEC prevention is so far largely lacking, making the peptidomic analysis of breast milk-derived exosomes appealing. In our study, a total of 719 peptides from milk exosomes were identified. Of these, 70 significant differences in peptide profiles based on term and preterm were determined. Many of the exosomal peptide precursors from both term and preterm milk are associated with metabolic process, developmental process, immune system process, biological adhesion, and cell proliferation. Our results confirm previous studies that demonstrate that milk exosomes have the potential to influence the immune response and growth.29 For example, milk exosomes can inhibit cytokine production from PBMCs and increase the number of T regulatory cells.30 As we know, a peptide and its precursor protein always have similar function. Moreover, the amino acid sequence within the protein domain region plays an important role in protein function. Therefore, by combining the GO analysis and functional domain analysis of differentially expressed peptides, we propose three upregulated peptides derived from protein domain regions (two derived from lactotransferrin (LTF) and one derived from lactadherin (MFGE8)) with a likelihood of proliferation and migration promoting functionality. Lactoferrin (LTF) is secreted by epithelial cells and abundantly exists in milk and other body fluids.31 LTF is well known for its anti-inflammatory and antimicrobial properties, as well as its roles in immunomodulation.32 LTF has recently been found to promote cell proliferation in various cell lines. Nichols et al. reported that LTF from milk stimulated rat intestinal cell proliferation.33 Tang et al. demonstrated that LTF not only significantly promoted fibroblast proliferation but also stimulated fibroblast migration as well as prevented fibroblasts from cell death.34 Lactadherin, also known as milk fat globule-EGF factor VIII (MFGE8), is one of the common milk proteins that is highly enriched in Preterm-Exos. It has been reported that MFGE8 could promote neural stem cell proliferation and migration in an αvβ3-integrin dependent manner.35 MFGE8 could also promote the regeneration of injured intestinal mucosa by accelerating migration and proliferation through protein kinase C-dependent pathway.36 Thus, we suggested that the two peptides derived from the Transferrin-like 1 domain of LTF and the peptide derived from the EGF-like domain of MFGE8 may have potential protective effects against NEC in neonates. Future studies need to further clarify the roles of these peptides.
To our knowledge, casein and casein-derived peptides have bioactive and regulatory functions in various intestinal diseases, such as protection of intestinal epithelial barrier function.37-39 In addition, a β-casein peptide (1-28) has been shown to enhance proliferation and IL-6 production of CD19+ cells by the TLR4-dependent pathway.40 The peptides produced from Alpha-S1-casein are considered to possess antioxidative and intestinal antiinflammatory activities.41 In the present study, we found that the altered peptide sequences were predominantly derived from the parent proteins β-casein and Alpha-S1-casein. Therefore, the potential function of these exosomal casein derived peptides (Figure S1, Supporting Information) needs to be investigated in the future.
Taken together, our study is the first report on the different peptidomic profiles between term and preterm milk exosomes. As described in this study, the peptides that are highly enriched in preterm milk exosomes may possess protective activities against NEC by promoting intestinal cell proliferation and migration. Since exosome is a new type of regulator in milk that has promising application values, our results will facilitate the goal of the prevention of NEC, as well as provide better nutrition for newborns.
Acknowledgements
X.W., X.Y, and L.Z. contributed equally to this work. X.Y.W., X.Y.Y., L.Z., J.Y.C., Y.H.Z., H.L., Y.H., W.J.C., S.L.X., B.B.L., T.C., and J.Z. collected and analyzed the data. Y.C., Z.B.Y., and S.P.H. designed the study and wrote the manuscript. All the authors approved the final submission. National Natural Science Foundation of China (81370200, 81701491, 81741089, 81601333, 81401232, 81501296); The Natural Science Foundation of Jiangsu Province, China (BK20170152, BK20181089, BK20141077); Jiangsu Province Women and Children Health Key Talents (FRC201740); Nanjing Medical Science and Technique Development Foundation (201605052); Nanjing Medical Science and Technology Development Foundation (ZKX16062); Jiangsu Women and Children Health Research Project (F201642); Medical Innovation Team of Jiangsu Province (CXTDB2017016), Wuxi Medical Development Discipline (FZXK001); Wuxi Key Medical Disciplines (ZDXK012); Wuxi Young Medical Talents (QNRC094); Wuxi Maternal and Child Health Research Project (FYKY201507).
Conflict of Interest
The authors declare no conflict of interest.




