Inhibitor Assessment against the LpxC Enzyme of Antibiotic-resistant Acinetobacter baumannii Using Virtual Screening, Dynamics Simulation, and in vitro Assays
Manel Zoghlami
Laboratoire de Mycologie, Pathologies et Biomarqueurs (LR16ES05), Département de Biologie, Université de Tunis-El Manar, 2092 Tunis, Tunisia
Search for more papers by this authorMaroua Oueslati
Laboratoire de Mycologie, Pathologies et Biomarqueurs (LR16ES05), Département de Biologie, Université de Tunis-El Manar, 2092 Tunis, Tunisia
Search for more papers by this authorZarrin Basharat
Jamil-ur-Rahman Center for Genome Research, Dr. Panjwani Center for Molecular Medicine and Drug Research, ICCBS University of Karachi, 75270 Karachi, Pakistan
Search for more papers by this authorNajla Sadfi-Zouaoui
Laboratoire de Mycologie, Pathologies et Biomarqueurs (LR16ES05), Département de Biologie, Université de Tunis-El Manar, 2092 Tunis, Tunisia
Search for more papers by this authorCorresponding Author
Abdelmonaem Messaoudi
- [email protected]
- /fax:0021624321186
Laboratoire de Mycologie, Pathologies et Biomarqueurs (LR16ES05), Département de Biologie, Université de Tunis-El Manar, 2092 Tunis, Tunisia
Higher Institute of Biotechnology of Beja, Jendouba University, Habib Bourguiba Street, 9000 Beja, Tunisia
Search for more papers by this authorManel Zoghlami
Laboratoire de Mycologie, Pathologies et Biomarqueurs (LR16ES05), Département de Biologie, Université de Tunis-El Manar, 2092 Tunis, Tunisia
Search for more papers by this authorMaroua Oueslati
Laboratoire de Mycologie, Pathologies et Biomarqueurs (LR16ES05), Département de Biologie, Université de Tunis-El Manar, 2092 Tunis, Tunisia
Search for more papers by this authorZarrin Basharat
Jamil-ur-Rahman Center for Genome Research, Dr. Panjwani Center for Molecular Medicine and Drug Research, ICCBS University of Karachi, 75270 Karachi, Pakistan
Search for more papers by this authorNajla Sadfi-Zouaoui
Laboratoire de Mycologie, Pathologies et Biomarqueurs (LR16ES05), Département de Biologie, Université de Tunis-El Manar, 2092 Tunis, Tunisia
Search for more papers by this authorCorresponding Author
Abdelmonaem Messaoudi
- [email protected]
- /fax:0021624321186
Laboratoire de Mycologie, Pathologies et Biomarqueurs (LR16ES05), Département de Biologie, Université de Tunis-El Manar, 2092 Tunis, Tunisia
Higher Institute of Biotechnology of Beja, Jendouba University, Habib Bourguiba Street, 9000 Beja, Tunisia
Search for more papers by this authorGraphical Abstract
Abstract
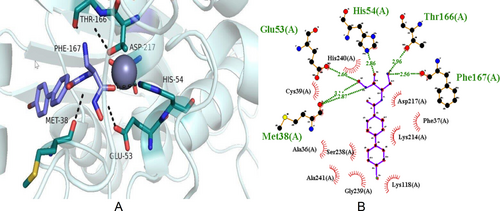
Background: Bacterial resistance is currently a significant global public health problem. Acinetobacter baumannii has been ranked in the list of the World Health Organization as the most critical and priority pathogen for which new antibiotics are urgently needed. In this context, computational methods play a central role in the modern drug discovery process. The purpose of the current study was to identify new potential therapeutic molecules to neutralize MDR A. baumannii bacteria. Methods: A total of 3686 proteins retrieved from the A. baumannii proteome were subjected to subtractive proteomic analysis to narrow down the spectrum of drug targets. The SWISS-MODEL server was used to perform a 3D homology model of the selected target protein. The SAVES server was used to evaluate the overall quality of the model. A dataset of 74500 analogues retrieved from the PubChem database was docked with LpxC using the AutoDock software. Results: In this study, we predicted a putative new inhibitor for the Lpxc enzyme of A. baumannii. The LpxC enzyme was selected as the most appropriate drug target for A. baumannii. According to the virtual screening results, N-[(2S)-3-amino-1-(hydroxyamino)-1-oxopropan-2-yl]-4-(4-bromophenyl) benzamide (CS250) could be a promising drug candidate targeting the LpxC enzyme. This molecule shows polar interactions with six amino acids and non-polar interactions with eight other residues. In vitro experimental validation was performed through the inhibition assay. Conclusion: To the best of our knowledge, this is the first study that suggests CS250 as a promising inhibitory molecule that can be exploited to target this gram-negative pathogen.
Conflict of interest
None declared.
Open Research
Data Availability Statement
Data available on request from the authors.
Supporting Information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
| Filename | Description |
|---|---|
| minf202200061-sup-0001-misc_information.pdf1.4 MB | Supporting Information |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
References
- 1M. Falagas, E. Karveli, The changing global epidemiology of Acinetobacter baumannii infections: a development with major public health implications, Vol.13, Elsevier, 2007, pp. 117–119.
- 2C. L. Holmes, M. T. Anderson, H. L. Mobley, M. A. Bachman, Clin. Microbiol. Rev. 2021, 34(2), e00234-20.
- 3B. Ingti, S. Upadhyay, M. Hazarika, A. B. Khyriem, D. Paul, P. Bhattacharya, S. Joshi, D. Bora, D. Dhar, A. Bhattacharjee, Res. Microbiol. 2020, 171, 128–133.
- 4G. Huys, M. Cnockaert, M. Vaneechoutte, N. Woodford, A. Nemec, L. Dijkshoorn, J. Swings, Res. Microbiol. 2005, 156, 348–355.
- 5S. Magnet, P. Courvalin, T. Lambert, Antimicrob. Agents Chemother. 2001, 45, 3375–3380.
- 6I. Kyriakidis, E. Vasileiou, Z. D. Pana, A. Tragiannidis, Pathogenesis 2021, 10, 373.
- 7W. Zhu, Y. Wang, W. Cao, J. Zhang, Jundishapur J Microbiol.. 2019, 12(6).
- 8X. Zhen, C. S. Lundborg, X. Sun, X. Hu, H. Dong, Antimicrob. Resist. Infect. Control 2019, 8(1), 1–23.
- 9B. Ingti, S. Upadhyay, M. Hazarika, A. B. Khyriem, D. Paul, P. Bhattacharya, S. Joshi, D. Bora, D. Dhar, A. Bhattacharjee, Res. Microbiol. 2020, 171, 128–133.
- 10E. Tacconelli, E. Carrara, A. Savoldi, S. Harbarth, M. Mendelson, D. L. Monnet, C. Pulcini, G. Kahlmeter, J. Kluytmans, Y. Carmeli, Lancet Infect. Dis. 2018, 18(3), 318–327.
- 11A. G. Dalecki, F. Wolschendorf, J. Microbiol. Methods 2016, 126, 30–34.
- 12N. Rahman, I. Muhammad, G. E. Nayab, H. Khan, R. Filosa, J. Xiao, S. T. Hassan, Curr. Top. Med. Chem. 2019, 19(29), 2708–2717.
- 13H. Kumar, F. Frischknecht, G. R. Mair, J. Gomes, Infect. Genet. Evol. 2015, 36, 72–81.
- 14H. Kumar, J. Kehrer, M. Singer, M. Reinig, J. M. Santos, G. R. Mair, F. Frischknecht, Expert Opin. Ther. Targets 2019, 23(3), 251–261.
- 15X. Xia, Curr. Top. Med. Chem. 2017, 17(15), 1709–1726.
- 16M. K. Yadav, S. Ahmad, K. Raza, S. Kumar, M. Eswaran, Km. M. Pasha, J. Biomol. Struct. Dyn. 2022, 1–13.
- 17P. Tiwari, P. Sharma, M. Kumar, A. Kapil, E. Abdul Samath, P. Kaur, J. Biomol. Struct. Dyn. 2021, 12, 1–16.
- 18V. Tiwari, M. Tiwari, D. Biswas, J.Antibiot. 2018, 71, 522–534.
- 19K. Malathi, S. Ramaiah, J. Cell. Biochem. 2019, 120(1), 584–591.
- 20M. M. Trush, V. Kovalishyn, D. Hodyna, O. V. Golovchenko, S. Chumachenko, I. V. Tetko, V. S. Brovarets, L. Metelytsia, Chem. Biol. Drug Des. 2020, 95(6), 624–630.
- 21K. Clark, I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, E. W. Sayers, Nucleic Acids Res. 2016, 44(D1), D67–D72.
- 22W. Li, L. Jaroszewski, A. Godzik, Bioinformatics. 2001, 17(3), 282–283.
- 23R. Zhang, H. Y. Ou, C. T. Zhang, Nucleic Acids Res. 2004, 32(suppl_1), D271–D272.
- 24S. F. Altschul, W. Gish, W. Miller, E. W. Myers, D. J. Lipman, J. Mol. Biol. 1990, 215(3), 403–410.
- 25M. Goyal, S. Citu, N. Singh, Asian J. Pharm. Clin. Res. 2018, 11, 230–236.
10.22159/ajpcr.2018.v11i3.22105 Google Scholar
- 26G. Lubec, L. Afjehi-Sadat, J. W. Yang, J. P. P. John, Prog. Neurobiol. 2005, 77(1–2), 90–127.
- 27G. S. Kumar, S. Sarita, G. M. Kumar, K. Pant, P. Seth, J. Antivir. Antiretrovir. 2010, 2, 038–041.
- 28M. Kanehisa, M. Furumichi, M. Tanabe, Y. Sato, K. Morishima, Nucleic Acids Res. 2017, 45(D1), D353–D361.
- 29N. Y. Yu, J. R. Wagner, M. R. Laird, G. Melli, S. Rey, R. Lo, P. Dao, S. C. Sahinalp, M. Ester, L. J. Foster, Bioinformatics. 2010, 26(13), 1608–1615.
- 30C. S. Yu, Y. C. Chen, C. H. Lu, J. K. Hwang, Proteins Struct. Funct. Bioinf. 2006, 64(3), 643–651.
- 31B. Bjellqvist, G. Hughes, Ch. Pasquali, N. Paquet, F. Ravier, J. Ch. Sanchez, S. Frutiger, D. F. Hochstrasser, Electrophoresis. 1993, 14, 1023–1031.
- 32T. Liu, Y. Lin, X. Wen, R. N. Jorissen, M. K. Gilson, Nucleic Acids Res. 2007, 35(suppl_1), D198–D201.
- 33D. S. Wishart, Y. D. Feunang, A. C. Guo, E. J. Lo, A. Marcu, J. R. Grant, T. Sajed, D. Johnson, C. Li, Z. Sayeeda, Nucleic Acids Res. 2018, 46(D1), D1074–D1082.
- 34H. M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, P. E. Bourne, Nucleic Acids Res. 2000, 28(1), 235–242.
- 35T. Schwede, J. Kopp, N. Guex, M. C. Peitsch, Nucleic Acids Res. 2003, 31(13), 3381–3385.
- 36G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell, A. J. Olson, J. Comput. Chem. 2009, 30(16), 2785–2791.
- 37S. Kim, J. Chen, T. Cheng, A. Gindulyte, J. He, S. He, Q. Li, B. A. Shoemaker, P. A. Thiessen, B. Yu, Nucleic Acids Res. 2021, 49(D1), D1388–D1395.
- 38E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, T. E. Ferrin, J. Comput. Chem. 2004, 25(13), 1605–1612.
- 39M. D. Eldridge, C. W. Murray, T. R. Auton, G. V. Paolini, R. P. Mee, J. Comput.-Aided Mol. Des. 1997, 11(5), 425–445.
- 40W. L. DeLano, The PyMOL molecular graphics system. http://www.pymol.org 2002.
- 41J. C. Abdul-Mutakabbir, N. C. Griffith, R. K. Shields, F. P. Tverdek, Z. K. Escobar, Infect. Dis. Ther. 2021, 10(4), 2177–2202.
- 42F. Pasteran, J. Cedano, M. Baez, et al., Antibiotics 2021, 5, 2.
- 43W. F. Penwell, A. B. Shapiro, R. A. Giacob, et al., Antimicrob. Agents Chemother. 2015, 3, 1680–9.
- 44G. L. Wood, J. A. Washington, in Manual of Clinical Microbiology (Ed.: P. R. Murray), ASM, Washington DC, 1995, pp. 1327–1341.
- 45F. Shahid, M. Shehroz, T. Zaheer, A. Ali, Front. Anti-infect. Drug Discov. 2020, 8, 144.
10.2174/9789811412387120080007 Google Scholar
- 46A. Hassan, A. Naz, A. Obaid, R. Z. Paracha, K. Naz, F. M. Awan, S. A. Muhmmad, H. A. Janjua, J. Ahmad, A. Ali, BMC Genomics 2016, 17(1), 1–25.
- 47A. P. Chan, G. Sutton, J. DePew, R. Krishnakumar, Y. Choi, X. Z. Huang, E. Beck, D. M. Harkins, M. Kim, E. P. Lesho, GenomeBiology 2015, 16(1), 1–28.
- 48S. Maurya, S. Akhtar, M. H. Siddiqui, M. K. A. Khan, Int Res J Eng Technol. 2020, 9, 262–273.
- 49K. E. Mdluli, P. R. Witte, T. Kline, A. W. Barb, A. L. Erwin, B. E. Mansfield, A. L. McClerren, M. C. Pirrung, L. N. Tumey, P. Warrener, Antimicrob. Agents Chemother. 2006, 50(6), 2178–2184.
- 50A. N. Sarangi, R. Aggarwal, Q. Rahman, N. Trivedi, J. comput. sci. syst. biol.. 2009, 2(5), 255–258.
- 51K. M. Krause, C. M. Haglund, C. Hebner, A. W. Serio, G. Lee, V. Nieto, F. Cohen, T. R. Kane, T. D. Machajewski, D. Hildebrandt, Antimicrob. Agents Chemother. 2019, 63(11), e00977–19.
- 52S. Ahmad, A. Navid, A. S. Akhtar, S. S. Azam, A. Wadood, INTERDISCIP SCI. 2019, 11(3), 508–526.
- 53J. Zhang, A. Chan, B. Lippa, J. B. Cross, C. Liu, N. Yin, J. A. C. Romero, J. Lawrence, R. Heney, P. Herradura, Bioorg. Med. Chem. Lett. 2017, 27(8), 1670–1680.
- 54C. J. Lee, X. Liang, R. Gopalaswamy, J. Najeeb, E. D. Ark, E. J. Toone, P. Zhou, ACS Chem. Biol. 2014, 9(1), 237–246.
- 55D. Pradhan, V. Priyadarshini, M. Munikumar, S. Swargam, A. Umamaheswari, A. Bitla, J. Biomol. Struct. Dyn. 2014, 32(2), 171–185.
- 56P. Zhou, A. W. Barb, Curr. Pharm. Biotechnol. 2008, 9(1), 9–15.
- 57A. P. Tomaras, C. J. McPherson, M. Kuhn, A. Carifa, L. Mullins, D. George, C. Desbonnet, T. M. Eidem, J. I. Montgomery, M. F. Brown, mBio 2014, 5(5), e01551–14.
- 58S. Skariyachan, N. Taskeen, M. Ganta, J. Biomol. Struct. Dyn. 2018, 37, 1146–1169.
- 59S. Skariyachan, N. Taskeen, M. Ganta, B. Krishna, Crit. Rev. Microbiol. 2019, 1040–841X, 1549–7828.
- 60C. Lee, J. H. Lee, M. Park, K. S. Park, K. I. I. Bae, Y. B. Kim, C. Cha, B. C. Jeong, S. H. Lee, Front. Cell. Infect. Microbiol. 2017, 7, 55.