Diagnosis of Menke-Hennekam syndrome by prenatal whole exome sequencing and review of prenatal signs
Abstract
Introduction
CREBBP truncating mutations and deletions are responsible for the well-known Rubinstein-Taybi syndrome. Recently, a new, distinct CREBBP-linked syndrome has been described: missense mutations located at the 3′ end of exon 30 and the 5′ portion of exon 31 induce Menke-Hennekam syndrome. Patients with this syndrome present a recognizable facial dysmorphism, intellectual disability of variable severity, microcephaly, short stature, autism, epilepsy, visual and hearing impairments, feeding problems, upper airway infections, scoliosis, and/or kyphosis. To date, all diagnoses were made postnatally.
Method and Case Report
Trio-whole exome sequencing (WES) was performed in a fetus showing increased nuchal translucency persistence and aorta abnormalities at 28 weeks of gestation (WG).
Results
WES revealed a CREBBP de novo missense mutation (c.5602C>T; p.Arg1868Trp) in exon 31, previously reported as the cause of Menke-Hennekam syndrome. Termination of pregnancy was performed at 32 WG. We further reviewed the prenatal signs of Menke-Hennekam syndrome already reported. Among the 35 patients reported and diagnosed postnatally up to this day, 15 presented recognizable prenatal signs, the most frequent being intra-uterine growth retardation, brain, and cardiovascular anomalies.
Conclusion
Menke-Hennekam is a rare syndrome with unspecific, heterogeneous, and inconstant prenatal symptoms occurring most frequently with the c.5602C>T, p.(Arg1868Trp) mutation. Therefore, the prenatal diagnosis of Menke-Hennekam syndrome is only possible by molecular investigation. Moreover, this case report and review reinforce the importance of performing prenatal WES when unspecific signs are present on imaging.
1 INTRODUCTION
CREBBP (OMIM #600140) encodes the transcriptional co-activator CREB-binding protein, belonging to the type 3 family of lysine acetyltransferases (KAT3). CREBBP is involved in histone and non-histone protein modification, thereby regulating chromatin accessibility and transcription (Dutto et al., 2018).
Heterozygous loss of function variants and deletions of CREBBP are involved in the well-known Rubinstein-Taybi syndrome type 1 (RTS1, OMIM 180849) characterized by a typical facial dysmorphism, characteristic grimacing smile, microcephaly, broad thumbs and halluces, intellectual disability and postnatal growth retardation (Milani et al., 2015).
However since 2016 and the first 11 patients reported by Menke et al., 33 patients presented a different phenotype with missense CREBBP mutations located at the 3′ end of exon 30 and the 5′ portion of exon 31, in the ZZ and TAZ2 domain, respectively, have been reported (Angius et al., 2019; Banka et al., 2019; Menke et al., 2016, 2018; Nishi et al., 2022; Sima et al., 2022). In addition, 2 patients had variants located in the CREBBP homologous area close to the TAZ2 domain of EP300, a well-known gene also usually involved in RTS (Menke et al., 2018). The phenotype differs substantially from that in RTS patients; for instance, grimacing smile, broad halluces, and thumbs are absent. The facial dysmorphism is also not similar to RTS patient's, usually consisting of ptosis, telecanthi, short, and upslanted palpebral fissures, depressed nasal ridge, short nose, anteverted nares, short columella, and long philtrum. Other characteristics of this new syndrome included intellectual disability of variable severity, microcephaly, short stature, autism, visual and hearing impairments, feeding problems, epilepsy, upper airway infections, scoliosis, and/or kyphosis.
Despite the prenatal symptoms indicated in the 35 patients, none of them reported any prenatal diagnosis yet. Hereby, we describe the first prenatal diagnosis of a Menke-Hennekam syndrome by whole exome sequencing and review the prenatally accessible signs. Finally, we will be discussing about the opportunity of whole exome sequencing in the context of limited prenatal symptoms.
2 CASE REPORT
The patient was a 32-year-old woman in her sixth pregnancy. She had two previous miscarriages and one extra-uterine gestation. This non-related couple also had two healthy daughters. The familial history revealed two male nephews, one from the father and one from the mother with an increased nuchal translucency history during pregnancy without any developmental disorder at 7 years old. Her sister was pregnant with a fetus with a septal ventricular defect at the same time. The mother had an ear surgery during early childhood without functional sequelae and had slight dysmorphological traits.
First-trimester ultrasonography (US) revealed a 4.7 mm increased nuchal translucency. The next US at 15 weeks and 6 days showed a septal ventricular defect and a right aortic cross. The increased nuchal translucency was still present. The chromosomal microarray analysis on amniotic cells was normal. Moreover, prefrontal edema, facial dysmorphism, moderate right pleural liquid, and suspicion of pulmonary stenosis were added to the previous features on two ultrasounds performed at 20 and 24 weeks of gestation (WG).
The patient was then referred to our hospital, where an echocardiograph was performed at 25 WG. It revealed an unusual appearance of the aorta, with a contoured aspect at its junction with the ductus arteriosus, without ventricular septal defect. Also, this echocardiograph confirmed the persistence of the increased nuchal translucency and the prefrontal edema. Among the possible diagnoses, a rasopathy (Noonan syndrome) was mentioned. The couple was counseled at 28 WG about the uncertain prognosis and offered prenatal whole exome sequencing (WES) which was performed on parental DNA extracted from blood samples and fetal DNA extracted from amniotic cells.
Whole exome sequencing analysis revealed a CREBBP de novo missense mutation (c.5602C>T; p.Arg1868Trp) in exon 31. Arginine at position 1868 is a highly evolutionarily conserved amino acid across species, and in silico prediction tools showed a high pathogenic score of: 28, 0.99, and 0 for CADD, Polyphen-2, and SIFT, respectively. This variant is absent from gnomAD database and has been previously reported as the cause of Menke-Hennekam syndrome (Angius et al., 2019; Banka et al., 2019; Giles et al., 1998; Giordano & Avantaggiati, 1999; Janknecht & Hunter, 1996; Menke et al., 2016; Nishi et al., 2022; Richards et al., 2015; Sima et al., 2022; Vo & Goodman, 2001). It was therefore classified as a pathogenic variation according to ACMG criteria (Richards et al., 2015).
In light of the severity of the prognosis, termination of pregnancy was demanded by the couple and performed at 32 WG in accordance with French law. Parents declined post-mortem examination.
3 RESULTS AND DISCUSSION
Cyclic-AMP response element binding protein (CREBBP) is a large transcriptional adaptor protein that interacts with a variety of cellular transcription factors, thus mediating transcriptional responses to numerous intra and extracellular signals (Giordano & Avantaggiati, 1999; Janknecht & Hunter, 1996; Vo & Goodman, 2001). For instance, it plays an important role in DNA damage response: after DNA damage, the histone acetylation mediated by CREBBP enables transcription activation and facilitates the recruitment of DNA repair factors to the damaged site.
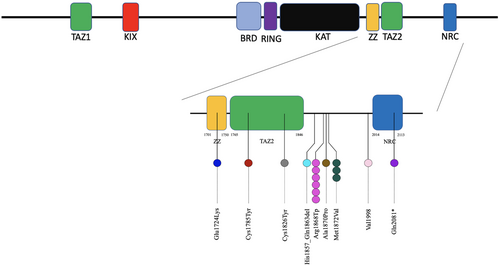
CREBBP is composed of structural and functional domains that mediate the interactions with other proteins (Giles et al., 1998; Janknecht & Hunter, 1996). Three putative zinc-binding domains were originally termed CH1, CH2, and CH3 by Borrow et al. (1996) on the basis of the sequence positions of cysteine and histidine residues. Ponting et al. suggested an alternative nomenclature where CH1 corresponds to TAZ1, CH2 region to a PHD zinc finger motif, and the CH3 domain incorporates two zinc-binding motifs, ZZ (Zinc finger, ZZ-type; residues 1701 to 1744) and TAZ2 (Zinc finger, TAZ type; residues 1765 to 1846). The role of these domains is to stabilize helical folding and mediate interactions with transcriptional regulatory proteins by mediating Zn2+ binding (De Guzman et al., 2000; Ponting et al., 1996).
All variants responsible for Menke-Hennekam syndrome are missense variants located on exon 30/31 overlapping the ZZ and TAZ2 domains which contrasts with the wide distribution of RTS variants, spreading through the entire gene, which are mainly truncating variants. These variant clusters are predicted to perturb the proper folding and stability of the structural organization of the zinc finger domains, thus affecting their binding properties (De Guzman et al., 2000; Legge et al., 2004). Seven of the 35 patients yet reported had the same c.5602C>T p.(Arg1868Trp) variant localized close to TAZ2, in the c-terminal part of CREBBP: Patients #9 (4 years old female) and #10 (9 and a half months old female) of the first report (Menke et al., 2016); C17 (2 years old male), C18 (4 years old male), C19 (1-year-old female)of the next report (Menke et al., 2018), an additional 20 months girl (Patient #2) and 2-month-old male, more recently described (Banka et al., 2019).
Globally, prenatal history was unremarkable in most of Menke-Hennekam patients reported, even if prenatal detectable signs were present in 15/35 (43%) patients (Table 1, Figure 1). Among patients with prenatal detectable signs, four out of seven (57%) carried the same CREBBP c.5602C>T p.(Arg1868Trp) mutation, suggesting a higher prevalence of prenatal manifestations. The main symptoms discovered prenatally were fetal growth, weight or skull circumference restriction (10/15), microcephaly (5/15), cardiac septal defect (5/15), cleft palate (4/15), corpus callosum abnormalities and bilateral hydronephrosis (2/15). A large range of symptoms from abnormal extremities (pre and postaxial polydactyly) to brain malformations/defects (ventriculomegaly), all found once in each patient, completed the spectrum. Interestingly, among the two patients with EP300 variants, neither had prenatal signs meaning that all eight patients were only CREBBP-mutated. Of note, IUGR and microcephaly in patient #3 from Banka et al. could be linked to the maternal CMV infection and not to the CREBBP mutation.
| Patient | A1 | B3 | M5 | N1 | M10 | M17 | M18 | B2 | S1 | Case report | M21 | M22 | N2 | N3 | N4 | N6 | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variant | c.5170G>A p.(Glu1724Lys) | c.5354G>A p.(Cys1785Tyr) | c.5478 p.(Cys1826Trp) | c.5570_5590del p.(His1857_Gln 1863del) | c.5602C>T p.(Arg1868Trp) | c.5602C>T p.(Arg1868Trp) | c.5602C>T p.(Arg1868Trp) | c.5602C>T p.(Arg1868Trp) | c.5602C>T p.(Arg1868Trp) | c.5602C>T p.(Arg1868Trp) | c.5608G>C p.(Ala1870Pro) | c.5614A>G p.(Met1872Val) | c.5614A>G p.(Met1872Val) | c.5614A>G p.(Met1872Val) | c.5991delC p.(Val1998) | c.6241C>T p.(Gln2081*) | |
| Intra-uterine growth retardation | − | + | + | + | + | − | − | + | + | − | − | − | + | + | + | + | 10/36 |
| Nuchal translucency | − | − | − | − | − | − | + | − | − | + | − | − | − | − | − | − | 2/36 |
| Oligohydramnios | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | 1/36 |
| Cleft palate | + | − | − | − | − | − | + | − | + | − | + | − | − | − | − | − | 4/36 |
| Polydactyly | − | − | − | − | − | Pre axial | − | − | − | − | − | − | − | − | − | Post axial | 2/36 |
| Bilateral hydronephrosis | − | − | − | − | − | − | − | + | − | − | + | − | − | − | − | − | 2/36 |
| Cardiovascular anomalies | 5/36 | ||||||||||||||||
| Atrial septal defect | − | − | − | − | − | − | − | − | + | − | + | − | − | + | − | − | 3/36 |
| Ventricular septal defect | − | − | − | − | − | − | − | + | − | − | − | + | − | − | − | − | 2/36 |
| Pulmonary stenosis | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | 1/36 |
| Brain anomalies | 7/36 | ||||||||||||||||
| Microcephaly | − | + | + | − | + | − | − | + | + | + | − | − | − | − | − | − | 6/36 |
| Corpus callosum anomaly | − | − | − | − | − | − | − | + | − | − | − | + | − | − | − | − | 2/36 |
| Ventriculomegaly | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | 1/36 |
| Neuromuscular | 2/36 | ||||||||||||||||
| Decreased fetal movement | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | 1/36 |
| Multiple congenital contractures | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | 1/36 |
- Note: B2 and B3: Patient #2 and Patient #3 from Banka et al. (2019); M5 and M10: Patient 5 and Patient 10 from Menke et al. (2016); M17, M18, M21, and M22: Case 17, Case 18, Case 21 and Case 22 from Menke et al. (2018); A1: case report from Angius et al. (2019); N1, N2, N3, N4, N6: Patient 1, Patient 2, Patient 3, Patient 4, and Patient 6 from Nishi et al. (2022); S1: case report from Sima et al. (2022).
The unspecificity and heterogeneity of prenatal signs of Menke-Hennekam syndrome lead to the conclusion that the diagnosis is only possible by molecular investigation prenatally and highlight the importance of performing prenatal WES when unspecific signs are present on imaging. As far as genetic counseling is concerned, given the postnatal severity of this syndrome consisting of intellectual deficiency, epilepsy, seizures, visual, and hearing impairments, termination of pregnancy might be an option for the couples, depending on the maternal current pregnancy state law. The genetic result also allows accurate genetic counseling to the couple, the recurrence risk for their future pregnancies being reduced to the one of germline mosaicism.
In conclusion, Menke-Hennekam is a rare syndrome with heterogeneous and inconstant prenatal symptoms occurring most frequently with the c.5602C>T, p.(Arg1868Trp) mutation. When present, the most frequent signs are IUGR, microcephaly, cleft palate, and atrial septal defect. It can also be added to the list of syndromes with apparently isolated persistent nuchal translucency.
4 METHODS
After genetic counseling signed consents from both parents were obtained for the genetic analysis. Trio prenatal exome sequencing was performed in a diagnostic setting on parental DNA extracted from whole blood samples and fetal DNA extracted from the amniotic cells.
Library generation, exome enrichment, and sequencing were performed at the Genomic Platform of University hospital Necker-Enfants-Malades (APHP Paris, France). Libraries were prepared from 50 ng of fragmented genomic DNA. Exome enrichment was performed using Twist Human RefSeq Exome Kit, 36 Mb (Twist Bioscience kits) according to the manufacturer's recommendations, with DNA multiplexing by molecular barcoding for sample traceability. DNA libraries were then sequenced on Illumina NextSeq according to the manufacturer's recommendations. Bioinformatics analysis was performed by the Bioinformatics platform of the University of Paris Cité and Imagine Institute. Briefly, after demultiplexing, sequence reads were aligned to the human genome (NCBI build37/hg19) using BWA software. Downstream processing was carried out with the Genome analysis toolkit GATK (Haplotypecaller, Unifigenotyper, Samtools, and Freebayes). All variants with a read coverage ≤5× and a Phred-scaled quality of ≤20 were filtered out. Search for deletions and exonic duplications is performed by comparison of reading depths after normalization. Variants were annotated and filtered using a local interface (PolyWeb) developed by the Bioinformatics platform according to the current HGVS nomenclature, assisted by tools for databases (gnomAD, HGMD-Pro, Clinvar) and prediction software (Polyphen-2 HumanVar (PPN2), SIFT, CADD, Mutation Taster, MaxEntScan, SSF-like, NNSplice, and Gene Splicer). Variants with a frequency >1% in GnomAD were filtered out from the analysis. The analysis mainly focused on frameshift, nonsense, mature miRNA, start/stop codon loss, splice acceptor/donor site, missense variants with rare frequency (<1% in dbSNP, the 1000 Genomes Project, GnomAD, Exome Aggregation Consortium, and Exome Variant Server or variants identified more than 80 times in local sequencing data). Variants were classified according to the ACMG classification criteria (Richards et al., 2015).
AUTHOR CONTRIBUTIONS
Guillaume Cogan: He participated in the genetic diagnosis, discussions about the pathogenicity of the variant, the prognosis of the future child, and the delivery of the genetic counseling. The main writer of the paper, built up Figure 1 and Table 1, and performed most of the literature review and data-gathering about prenatal signs of Menke-Hennekam syndrome. Nicolas Bourgon and Julien Stiernemann : performed regular prenatal ultrasounds contributed to the genetic counseling, and the patient decision of pregnancy termination. They participated in the writing and reviewing of the paper, especially the ultrasonography description section. Roxana Borghese: was highly involved in discussions about genetic variant severity and participated in diagnosis announcement and genetic counseling. About the article, she participated in reviewing, especially the genetic counseling part. Emmanuel Julien: he performed the first pregnancy ultrasounds before her transfer to Necker-Enfants-Malades hospital, and contributed to the description of the first ultrasonography images. Aurélia Jacquette:participated in the discussion about the severity of the genetic variant and the genetic counseling to the couple and actively participated in the writing and reviewing. Bertrand Stos: Performed the echocardiography and the description of the echocardiography pictures. Amale Achaiaa and Sophie Chuon performed the DNA extraction, library preparation, and sequencing of the trio. They participated in the reviewing of the paper, especially on the technical part of the method. Patrick Nitschke, head of the Bioinformatic department of the imagine institute, developed the local interface “Polyweb” used by the laboratory for the analysis of genetic variants. He actively participated in the reviewing of the paper, mainly in the bioinformatic part of the method section. Cécile Fourrage:is the bioinformatician, who treated the sequencing data. She actively participated in the writing and reviewing of the paper, mainly in the bioinformatic part of the method section. Lucile Boutaud molecular geneticist actively interpretated the exome data and edition of the result She also actively wrote, annotated, and gave opinions about the article. Tania Attie-Bitach geneticist of the prenatal diagnosis department was involved in all the discussions about the patient, from the opportunity to perform a genetic analysis to the delivery of the diagnosis, the discussion of the pathogenicity, and the genetic counseling. She actively participated and coordinated the redaction of the present article.
ACKNOWLEDGMENTS
The authors thank the patient's family for participating in this study.
FUNDING INFORMATION
The author(s) received no specific funding for this work.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
Data supporting the findings from this study are available from the corresponding author on request.




