Identification of nine novel variants across PAX3, SOX10, EDNRB, and MITF genes in Waardenburg syndrome with next-generation sequencing
Abstract
Background
Waardenburg syndrome (WS) is a hereditary, genetically heterogeneous disorder characterized by variable presentations of sensorineural hearing impairment and pigmentation anomalies. This study aimed to investigate the clinical features of WS in detail and determine the genetic causes of patients with clinically suspected WS.
Methods
A total of 24 patients from 21 Han-Taiwanese families were enrolled and underwent comprehensive physical and audiological examinations. We applied targeted next-generation sequencing (NGS) to investigate the potential causative variants in these patients and further validated the candidate variants through Sanger sequencing.
Results
We identified 19 causative variants of WS in our cohort. Of these variants, nine were novel and discovered in PAX3, SOX10, EDNRB, and MITF genes, including missense, nonsense, deletion, and splice site variants. Several patients presented with skeletal deformities, hypotonia, megacolon, and neurological disorders that were rarely seen in WS.
Conclusion
This study revealed highly phenotypic variability in Taiwanese WS patients and demonstrated that targeted NGS allowed us to clarify the genetic diagnosis and extend the genetic variant spectrum of WS.
1 INTRODUCTION
Waardenburg syndrome (WS) is a hereditary auditory-pigmentary disorder characterized by sensorineural hearing impairment (SNHI) and pigmentation abnormalities of the eye, hair, and skin (Bondurand et al., 2007). The disease has an estimated prevalence rate of 1/42,000(Read & Newton, 1997) and accounts for 2%–5% of patients with congenital hearing impairment (Zaman et al., 2015).
WS is classified into four subtypes according to the presented clinical phenotype. Type 1 WS (WS1, OMIM#193500) is diagnosed according to the criteria proposed by the Waardenburg Consortium (Farrer et al., 1992) that states that a patient must have two major criteria or one major plus two minor criteria to be considered as affected. Of all the symptoms of WS, dystopia canthorum is the most distinctive feature of WS1 (Read & Newton, 1997). Type 2 WS (WS2) is distinguished from WS1 by the absence of dystopia canthorum (Iso et al., 2008). Based on its genetic heterogeneity, WS2 is further divided into WS2A (OMIM#193510), WS2B (OMIM% 600,193), WS2C (OMIM%606,662), WS2D (OMIM#608890), and WS2E (OMIM#611584). Type 3 WS (WS3, OMIM#148820), also called Klein–Waardenburg syndrome, presents as musculoskeletal abnormalities with the clinical features of WS1(Tekin et al., 2001). Type 4 WS (WS4), named Shah–Waardenburg syndrome, has similar traits to WS2 but is accompanied by Hirschsprung disease (HD, OMIM#142623) (Ahmed jan et al., 2021). WS4 is additionally classified into three subgroups, WS4A (OMIM#277580), WS4B (OMIM#613265), and WS4C (OMIM#613266), according to genetic characteristics.
WS presents with a high degree of genetic heterogeneity. Six genes have been confirmed to cause WS: paired box 3 (PAX3, OMIM*606597) (Boudjadi et al., 2018), microphthalmia-associated transcription factor (MITF, OMIM*156845) (Lautenschlager et al., 1996; Yang, Li, et al., 2013), SRY-box transcription factor 10 (SOX10, OMIM* 602229) (Iso et al., 2008; Yu et al., 2020), endothelin 3 (EDN3, OMIM*131242) (Pingault et al., 2001), endothelin receptor type B (EDNRB, OMIM*131244) (Issa et al., 2017), and snail family transcriptional repressor 2 (SNAI2, OMIM*602150) (Otręba et al., 2013). Variants in PAX3 are primarily responsible for WS1 and WS3, while SOX10, EDN3, and EDNRB variants are involved in WS4. Genetic studies have linked WS2 to variants in MITF, SOX10, EDN3, EDNRB, and SNAI2 (Huang et al., 2021). Interactions between the six WS-associated genes are believed to form a MITF-centered regulatory framework responsible for controlling the differentiation and development of neural crest cells (NCCs), particularly melanocytes derived from NCCs (Hou & Pavan, 2008; Huang et al., 2021). An abnormality in this framework could lead to the development of WS and related diseases (Hou & Pavan, 2008).
WS patients display phenotypic variability and reduced penetrance, making it difficult to diagnose the disease solely on the basis of clinical symptoms (Pingault et al., 2010). Additionally, although more than 400 pathogenic variants have been identified to cause WS (Zhang et al., 2021), there are still a number of cases that are unexplained at the molecular level (Pingault et al., 2010). As such, the identification of novel variants will contribute to a better understanding of WS pathogenesis. Meanwhile, phenotypic analysis of WS patients is also crucial for better determining the correlations between genotypes and phenotypes for each WS subtype, in turn providing guidance for genetic counseling, diagnosis, and treatment choices for patients (Huang et al., 2021). In this study, we examined in detail the clinical phenotypes in a cohort of individuals with suspected WS and performed comprehensive genetic analyses using a targeted next-generation sequencing (NGS) approach.
2 METHODS
2.1 Study patients
This study included 24 patients with clinically suspected WS from 21 unrelated Han-Taiwanese families. The diagnoses were made by experienced otolaryngologists or pediatricians based on the presentation of at least one major diagnostic criterion of the syndrome (Farrer et al., 1992): SNHI, blue iris, white forelock, or dystopia canthorum. Fourteen individuals with suspected WS were patients at National Taiwan University Hospital, whereas ten had been patients at ten other hospitals and were referred to National Taiwan University Hospital for genetic testing.
2.2 Characterization of phenotypes
We obtained and analyzed the family history, medical records, and results of physical, dermatological, musculoskeletal, neurologic, and audiological examinations of all participants. Additionally, the patients underwent otoscopic and audiometric evaluations performed by experienced audiologists using pure tone audiometry or diagnostic auditory brainstem response, depending on the patient's age and neurologic status (Lin et al., 2017; Newton, 2002).
2.3 Next-Generation sequencing
We collected peripheral blood from the patients to extract genomic DNA from mononuclear cells. A sonication method (Covaris, Woburn, MA) was used to produce DNA fragments of an average size of 800 bp, and the length and concentration of the fragments were measured using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) and Qubit (Thermo Scientific, Waltham, MA), respectively. A DNA library was constructed from the fragments using a TruSeq Library Preparation Kit (Illumina Inc., San Diego, CA). Probe capture-based target enrichment was achieved with a SeqCap EZ Hybridization and Wash Kit (Roche NimbleGen, Madison, WI) using probes designed to target the coding regions of 30 common deafness-associated genes in the Taiwanese population, including the six genes (PAX3 (NM_181459.4), MITF (NM_198159.3), EDNRB (NM_001201397.1), EDN3 (NM_003068.5), SOX10 (NM_006941.4), SNAI2 (NM_003068.5)) reported to be causative of WS (Table S1) (Huang et al., 2021; Wu et al., 2019). The total target region size was approximately 317 kb, and the DNA samples were sequenced with a MiSeq platform (Illumina Inc., San Diego, CA) to produce 300-nucleotide paired-end reads with a 150x average read depth.
2.4 Data analyses
The paired-end reads from the NGS sequencing were aligned, sorted, and converted using the BWA-MEM and Sort utility in the Sentieon DNAseq pipeline version 2018 (https://www.sentieon.com/products/) (Kendig et al., 2019). The Sentieon Haplotyper algorithm was used to detect variants, including single nucleotide substitutions and small insertions and deletions. We used ANNOVAR version 2019 (https://annovar.openbioinformatics.org/en/latest/#annovar-documentation) (Wang et al., 2010) to annotate all of the called variants with a series of information, including Human Genome Variation Society nomenclatures, maximum allele frequency across distinguished populations of the gnomAD database (Karczewski et al., 2020), minor allele frequency in the Taiwan Biobank database (Wei et al., 2021), and various in silico prediction outcomes, including PolyPhen-2 version 2 (Harvard University, Cambridge, MA), SIFT version 2019 (SANS Institute, North Bethesda, MD), LRT version 2009 (Washington University, St. Louis, MO), MutationTaster version 2 (Charité e Universitätsmedizin Berlin, Berlin, Germany), VariantAssessor version 3 (Computational Biology Center, Memorial Sloan-Kettering Cancer Center, NY), FATHMM version 2.3 (University of Bristol, Bristol, England), and MetaLR version 2015 (Human Genetics Center, University of Texas Health Science Center at Houston, Houston, TX). We excluded variants with an allele frequency of more than 1% in both the gnomAD and Taiwan Biobank database and chose the filtered variants that were within exome or intron splicing sites. Human disease databases, including ClinVar (Landrum et al., 2016) and Deafness Variation Database (Azaiez et al., 2018), were used to identify previously reported pathogenic variants. The online platform VarSome (Kopanos et al., 2019) was adopted to carefully assess all of the filtered variants in accordance with the American College of Medical Genetics and Genomics (ACMG) guidelines (Richards et al., 2015). The variants that met the criteria for pathogenic and likely pathogenic were recorded as disease-causing and confirmed with Sanger sequencing.
3 RESULTS
3.1 Clinical phenotypes
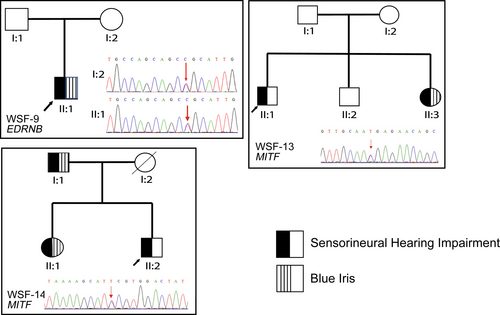
A total of 24 patients from 21 unrelated families were recruited for this study (14 female and 10 male patients). Four probands of the participants were clinically suspected of having WS1 (4/21, 19.0%) and seven of having WS2 (7/21, 33.3%); the other 10 probands had at least one major symptom of WS but failed to fulfill the criteria for a definite clinical diagnosis (Ahmed jan et al., 2021; Farrer et al., 1992). Genetic diagnoses were confirmed in 19 WS patients from 16 families (see “Variants Findings” below). Considering the penetrance of the causative variants, SNHI (13/19, 68.4%) and blue iris (13/19, 68.4%) were the most frequent phenotypes among these patients. White forelock was observed in two patients (2/19, 10.5%), and dystopia canthorum was observed in four patients (4/19, 21.1%). Three patients were members of the WSF-13 family, and two other patients belonged to the WSF-14 family (Figure 2). WSF-1 presented with dystopia canthorum and premature graying of hair, which was also observed in WSF-11. Nasolacrimal duct aplasia was discovered in WSF-2. We noted that WSF-6 exhibited hypotonia and inner ear deformity, confirmed by magnetic resonance imaging, and WSF-10 presented with megacolon. In addition, WSF-12 was reported to have facial dysmorphism and bilateral hydrocephalus. Table 1 lists the clinical features and affected genes of the 19 patients with confirmed genetic diagnoses.
| Subjects | Gender | SNHI | Blue iris | White forelock | Dystopia canthorum | Family historya | Other abnormality | WS types | Affected genes |
|---|---|---|---|---|---|---|---|---|---|
| WSF-1 | F | − | + | + | + | − | Synophrys, Broad high nasal root, PGH | WS1 | PAX3 |
| WSF-2 | F | − | + | − | + | N/Ab | Nasolacrimal duct aplasia | WS1 | PAX3 |
| WSF-3 | F | + | − | − | − | − | − | N/A | PAX3 |
| WSF-4 | F | + | − | − | − | − | − | N/A | PAX3 |
| WSF-5 | M | − | + | − | + | − | − | WS1 | PAX3 |
| WSF-17 | F | + | − | − | − | − | − | N/A | PAX3 |
| WSF-6 | F | + | + | − | − | − | Hypotonia, Inner ear deformityc | WS2 | SOX10 |
| WSF-7 | M | + | + | − | − | − | − | WS2 | SOX10 |
| WSF-8 | M | + | + | − | − | − | − | WS2 | SOX10 |
| WSF-9-II:1 | M | + | + | − | − | − | − | WS2 | EDNRB |
| WSF-10 | F | − | + | − | − | − | Megacolon, Brown hair | N/A | EDNRB |
| WSF-11 | F | − | + | − | + | − | Constipationd, PGH | WS1 | EDNRB |
| WSF-12 | F | + | − | + | − | − | Facial dysmorphism, Bilateral hydrocephalus | WS2 | EDNRB |
| WSF-13-I:1 | M | + | + | − | − | + | − | WS2 | MITF |
| WSF-13-II:1 | F | + | + | − | − | + | − | WS2 | MITF |
| WSF-13-II:2 | M | + | − | − | − | + | − | WS2 | MITF |
| WSF-14-II:1 | M | − | + | − | − | + | − | WS2 | MITF |
| WSF-14-II:3 | F | + | + | − | − | + | − | WS2 | MITF |
| WSF-15 | M | + | − | − | − | − | − | N/A | MITF |
- Abbreviations: F, female; M, male; N/A, not applicable to the diagnostic criteria for WS; PGH, premature graying of hair; SNHI, sensorineural hearing impairment; WS, Waardenburg syndrome.
- a First-degree relative previously diagnosed with WS.
- b WSF-2's mother had profound hearing loss but had not received genetic testing for WS.
- c WSF-6's magnetic resonant imaging showed bilateral cochlear deformity with reduced size and only 2 turns.
- d WSF-11's abdominal x-ray revealed no dilated bowel loop, which made it less likely to diagnose megacolon.
3.2 Variant findings
Causative gene variants of EDNRB, MITF, PAX3, or SOX10 were identified in 19 patients, with 14 of the variants being unique. Nine of the variants were novel variants and presented in 11 patients, and five previously reported variants were identified in eight patients (Table 2 and Table S2) (Bocángel et al., 2018; Chi, 2005; George et al., 2016; Lin et al., 2008; Richard et al., 2019; Wang et al., 2018; Yang, Dai, et al., 2013; Yang, Li, et al., 2013; Zhou et al., 2006). The novel variants included missense, nonsense, and splice site changes, as well as two frameshift deletions.
| Subjects | Variant profile | ACMG criteria | |||
|---|---|---|---|---|---|
| Variants | Structure | Protein change | Zygosityb | ||
| WSF-1 | NM_181459.4:c.807C>A, Missense | Exon | p.Asn269Lys | 0/1 | PS1, PM1, PM2, PP2, PP3 (P) |
| WSF-2 | NM_181459.4:c.52C>T, Nonsense | Exon | p.Gln18Ter | 0/1 | PVS1, PM2, PP3 (P) |
| WSF-3 | NM_181459.4:c.192C>A, Missense | Exon | p.His64Gln | 0/1 | PM1, PM2, PP2, PP3 (LP) |
| WSF-4 |
NC_000002.12(NM_013942.4)c: c.587-2A>G, Splice site |
Intron | N/A | 0/1 | PVS1, PM2 (LP) |
| WSF-5 | NM_181459.4:c.812G>A, Missense | Exon | p.Arg271His | 0/1 | PP5, PM1, PM2, PM5, PP2, PP3 (P) |
| WSF-17 | NM_181459.4: c.1130C>G, Missense | Exon | p.Ser377Cys | 0/1 | PM2, PP2, PP3 (LP) |
| WSF-6 | NM_006941.4:c.684_693del, Deletion | Exon | p.Glu229GlnfsTer54 | 0/1 | PVS1, PM2, PP3 (P) |
| WSF-7 | NM_006941.4:c.314_315del, Deletion | Exon | p.Lys105ThrfsTer28 | 0/1 | PVS1, PM2, PP3 (P) |
| WSF-8 | NM_006941.4:c.424T>G, Missense | Exon | p.Trp142Gly | 0/1 | PM1, PM2, PM5, PP2, PP3 (P) |
| WSF-9-II:1 | NC_000013.10(NM_001201397.1):c.754-2A>G, Splice site | Intron | N/A | 0/1 | PVS1, PM2, PP3 (P) |
| WSF-10 | NM_001201397.1:c.823G>A, Missense | Exon | p.Val275Met | 1/1 | PM1, PM2, PP3 (LP) |
| WSF-11 | NM_001201397.1:c.823G>A, Missense | Exon | p.Val275Met | 1/1 | PM1, PM2, PP3 (LP) |
| WSF-12 | NM_001201397.1:c.823G>A, Missense | Exon | p.Val275Met | 0/1 | PM1, PM2, PP3 (LP) |
| WSF-13-I:1 | NM_198159.3:c.1052C>T, Missense | Exon | p.Ser351Phe | 0/1 | PM1, PM2, PM5, PP2, PP3 (P) |
| WSF-13-II:1 | NM_198159.3:c.1052C>T, Missense | Exon | p.Ser351Phe | 0/1 | PM1, PM2, PM5, PP2, PP3 (P) |
| WSF-13-II:2 | NM_198159.3:c.1052C>T, Missense | Exon | p.Ser351Phe | 0/1 | PM1, PM2, PM5, PP2, PP3 (P) |
| WSF-14-II:1 | NM_198159.3:c.1078C>T, Nonsense | Exon | p.Arg360Ter | 0/1 | PVS1, PM2, PP3, PP5 (P) |
| WSF-14-II:3 | NM_198159.3:c.1078C>T, Nonsense | Exon | p.Arg360Ter | 0/1 | PVS1, PM2, PP3, PP5 (P) |
| WSF-15 | NM_198159.3:c.1078C>T, Nonsense | Exon | p.Arg356Ter | 0/1 | PVS1, PM2, PP3, PP5 (P) |
- Abbreviations: ACMG, American College of Medical Genetics and Genomics; LP, likely pathogenic; N/A, not applicable; P, pathogenic; US, uncertain significance.
- a For detailed information about the reporting history and allele frequency of the identified variants, please refer to the Table S2.
- b 0/1: heterozygous, 1/1: alternative homozygous.
- c Transcript NM_013942.4 was used to demonstrate a likely pathogenic variant in a splice site of PAX3.
Of the nine novel variants, three missense variants and one nonsense variant of PAX3 were detected: NM_181459.4:c.192C>A in the paired domain, c.807C>A in the homeodomain, c.1130C>G in the transactivation domain, and c.52C>T in exon 1. Three heterozygous variants in SOX10 were identified: NM_006941.4:c.314_315del and c.424T>G in the high mobility group (HMG) domain and a frameshift deletion c.684_693del between the HMG and K2 domains. Additionally, the EDNRB variant NC_000013.10 (NM_001201397.1):c.754-2A>G occurred in the splice site located 2 bp upstream of exon 3, and a novel MITF heterozygous missense variant NM_198159.3:c.1052C>T was identified, which could cause an amino acid replacement in the homeodomain of MITF.
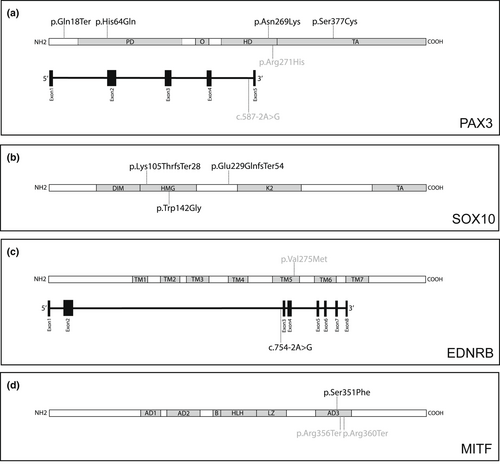
All five reported variants were either included in ClinVar (Landrum et al., 2016) or Deafness Variation Database (Azaiez et al., 2018), or both. The PAX3 heterozygous variant c.812G>A had been documented in 21 studies according to VarSome (Kopanos et al., 2019). Research about the EDNRB homozygous variant c.823G>A was reported in Taiwan (Lin et al., 2008), China (Zhou et al., 2006), and Pakistan (Richard et al., 2019). Moreover, in three Chinese studies (Wang et al., 2018; Yang, Dai, et al., 2013; Yang, Li, et al., 2013) and one Brazilian study (Bocángel et al., 2018), the MITF nonsense variant c.1066C>T was described. We confirmed each causative variant through Sanger sequencing. Table 2 and Figure 1 present the disease-causing variants identified in this study.
4 DISCUSSION
In this study, we identified 14 variants responsible for WS in 19 clinically suspected WS patients through targeted NGS (Table 2). All of these variants were pathogenic or likely pathogenic and located in the PAX3, SOX10, EDNRB, or MITF genes. Nine variants were novel. While the same missense variant EDNRB:c.823G>A, which was harbored by three unrelated patients (WSF-10, WSF-11, and WSF-12), had different pathogenicity according to ClinVar, Deafness Variation Database, and Varsome, we concluded that the variant might be disease-causing based on molecular analysis and previous case reports (discussed below).
PAX3 encodes a paired box transcription factor (TF) involved in the development of melanocytes through the regulation of MITF and other TFs (Galibert et al., 1999; Watanabe et al., 1998). Mice with heterozygous PAX3 variants might present with patchy loss of pigmentation, while those with homozygous variants die during pregnancy or shortly after birth (Pingault et al., 2010). In humans, PAX3 variants are the most prevalent cause of WS (Huang et al., 2021; Pingault et al., 2010) and are responsible for most WS1 and WS3 cases (Boudjadi et al., 2018; Huang et al., 2021). Homozygous or compound heterozygous PAX3 variants might induce more severe manifestations: extended depigmentation, upper limb defects, and even death in early infancy or in utero, for which WS3 is likely to be diagnosed (Mousty et al., 2015; Wollnik et al., 2003). In addition, the occurrence of de novo variants and germline mosaicism was found in some WS1 cases (Boudjadi et al., 2018). No apparent correlation between the variant type, location, or severity of the phenotype was observed in WS associated with PAX3 variants, suggesting that the pathophysiology of the disease may be due to a gene dosage effect (Pingault et al., 2010).
In this study, we identified four novel PAX3 variants. NM_181459.4:c.52C>T was located at exon 1 and might cause truncation of the translated protein. Variant c.192C>A was located at the paired domain (PD), which affects DNA binding capacity as well as the transcriptional regulation of downstream target genes (Pingault et al., 2010). The c.807C>A was located in the homeodomain (HD) and affects the interaction of PAX3 with DNA by destabilizing the domain fold (Birrane et al., 2009) (Figure 1a). The c. 1130C>G variant resided in the transactivation domain and may influence the transcription activity in cells (Boudjadi et al., 2018).
Our study also revealed that the six patients with PAX3 variants displayed diverse phenotypes. Aside from the major diagnostic criteria for WS, WSF-1 exhibited synophrys, broad high nasal roots, and premature graying of the hair. A similar finding to the nasolacrimal duct aplasia noted in WSF-2 was reported by David and Warin (David & Warin, 1972), wherein congenital blockage of the nasolacrimal duct was observed in an 11-year-old WS boy. More investigation is required to determine whether the abnormal development of the lacrimal duct is related to the dysmorphic effect that PAX3 variants may have on the craniofacial bone (Gad et al., 2008). In addition, WSF-3, WSF-4, and WSF-17 showed isolated SNHI, while WSF-5 had both blue iris and dystopia canthorum. These findings in our patients with PAX3 variants were consistent with the observation that the penetrance of each clinical feature of WS1 is not complete (Pingault et al., 2010), indicating that genetic backgrounds and environmental factors may influence the disease manifestations (Asher Jr. et al., 1996).
SOX10 encodes a TF critical to the early development of neural crest stem cells by promoting cell survival and maintaining multipotency (Kapur, 1999; Kelsh, 2006). Moreover, SOX10 controls the development of melanocytes by regulating MITF expression in synergy with PAX3 (Yu et al., 2020). Through modulation of RET (OMIM*164761) expression, SOX10 determines the morphogenesis of the enteric nervous system (ENS) (Bondurand et al., 2001). Mutant Sox10 heterozygous mice demonstrated variability of aganglionosis, the characteristic HD phenotype in WS4 patients (Southard-Smith et al., 1999). In humans, SOX10 variants account for ~15% of WS2 cases and 40–50% of WS4 patients (Bondurand et al., 2007). This study revealed three novel variants in SOX10. NM_006941.4:c.314_315del and c.424T>G were located in the highly conserved HMG domain of SOX10 that may cause conformational change and result in a decreased DNA-binding capacity (Hao et al., 2018). In addition, c.684_693del was identified in exon 3 of the gene, which activates nonsense-mediated RNA decay (NMD), leading to haploinsufficiency (Pingault et al., 2010) (Figure 1b).
SNHI was exhibited in all three patients with SOX10 variants. In a review of 417 WS patients (Song et al., 2016), SNHI was the most common feature of WS2 (100%) and WS4 (92.9%) in patients with SOX10 variants. Absent migration of NCC-derived melanocytes to the stria vascularis due to SOX10 variants might impair the ionic concentration of endolymph, which leads to the resultant SNHI (Syrris et al., 1999). Moreover, cochlear hypoplasia and the absence of the cochlear nerve were reported in patients with SOX10 variants (Barnett et al., 2009). However, we only found WSF-6 to have inner ear malformation of reduced cochlear size and turns. This finding demonstrates that while SOX10 variants might directly induce inner ear deformity (Hao et al., 2018), the phenotypic expression might be variable with incomplete penetrance, highlighting the difficulty of predicting inner ear malformations only with genotypic information.
Interestingly, besides typical manifestations of WS (SNHI and blue iris), WSF-6 also showed hypotonia without HD. Some WS4 patients with SOX10 variants were reported to have PCWH syndrome (OMIM#609136), a complex neurocristopathy including peripheral demyelinating neuropathy, central demyelinating leukodystrophy, Waardenburg syndrome, and Hirschsprung disease (Verheij et al., 2006). Patients with severe cases exhibit hypotonia, arthrogryposis, or respiratory insufficiency (Inoue et al., 2002; Touraine et al., 2000), while those with mild cases likely display variable hypotonia, spasticity, ataxia, and developmental delay (Pingault et al., 2000; Verheij et al., 2006). To date, several WS2 patients have also presented with neuromuscular features reminiscent of PCWH (Bondurand et al., 2007), thus delineating a new extended PCW phenotype (Pingault et al., 2010) similar to that observed in WSF-6.
EDNRB binds EDN3 and transmits signals through the Gαq/α11subunits of G-protein (Bondurand et al., 2018). The signaling pathway is crucial to NCC development during the embryonic stage of vertebrates, including melanoblast differentiation and the formation of neurons and glial cells in the ENS (Huang et al., 2021; Pingault et al., 2010). Mutant Ednr3 mice demonstrate hypopigmentation and the absence of enteric ganglia in the distal part of the gut (Bondurand et al., 2018; Touraine et al., 2000). The phenotypes observed in mutant Ednr3 mice were similar to those in WS4 patients, in whom homozygous EDNRB variants were first described (Attié et al., 1995). In contrast, heterozygous EDNRB variants were reported in patients with isolated HD (Amiel et al., 1996; Kusafuka et al., 1996). However, several studies have also proposed that the homozygous variants in EDNRB could induce isolated HD, and heterozygous variants might occur in WS4 (Pingault et al., 2001; Syrris et al., 1999). These findings suggest that EDNRB variants manifest in a more complex manner than merely as dominant or recessive conditions. Moreover, we might not be able to accurately forecast whether an EDNRB variant is more associated with isolated HD or WS because of the contribution of modifier genes to the development of both diseases (Bondurand et al., 2018).
In humans, EDNRB/EDN3 variants in WS2 patients make up less than 5% of cases and in WS4 patients, 20–30% of cases (Pingault et al., 2010). We identified four patients with EDNRB variants. WSF-9-II:1 had a novel heterozygous splice-site variant NC_000013.10(NM_001201397.1):c.754-2A>G two bp upstream of exon 3 (Figure 1c), which may lead to the absence of the genetic product (Richards et al., 2015). While WSF-9-II:1 was diagnosed with WS2, his mother, who harbored the same heterozygous variant, was not affected (Figure 2). WSF-10 and WSF-11 harbored the same homozygous c.823G>A variant (Table 2), which was initially identified in two Chinese patients and one Taiwanese individual in the heterozygous form with isolated HD (Lin et al., 2008; Zhou et al., 2006). However, WSF-10 and WSF-11 showed distinct phenotypes of WS. WS10 had megacolon and blue iris; WSF-11 exhibited blue iris, dystopia canthorum, premature graying of hair, and constipation (Table 1). Additionally, we found that although WSF-12 had heterozygous c.823G>A, she did not present with any gastrointestinal symptoms but possessed the phenotypes of WS2, facial dysmorphism, and bilateral hydrocephalus. Few cases of WS1 and WS4 were observed to have hydrocephalus, indicating WS increases the risk of neural tube defects in WS (Hart & Miriyala, 2017) and pathologic association with other developmental disorders (Yoder & Prayson, 2002). Clinically, WSF-10 did not meet the current diagnostic criteria of WS4 due to only presenting with blue iris and megacolon; WSF-11 was diagnosed as WS1, which is rarely caused by EDNRB variants (Cheng et al., 2019; Morimoto et al., 2018). These findings demonstrate the highly variable nature of phenotypic expression and the reduced penetrance of EDNRB features.
Additionally, we identified the missense variant c.823G>A located in transmembrane domain 5 (TM5, Figure 1c) (Huang et al., 2021) with different interpretations of pathogenicity in ClinVar (conflicting interpretation), DVD (pathogenic), and VarSome (likely pathogenic) (Table 2). The missense variants in TM5 of EDNRB were reported to cause dysfunctional intracellular signaling and impair the translocation of receptors to the plasma membrane (Fuchs et al., 2001; Tanaka et al., 1998). Together with the previous case reports (Lin et al., 2008; Richard et al., 2019; Zhou et al., 2006), these findings could lead to the interpretation of this variant to be disease-causing.
MITF is a basic helix–loop–helix zipper (bHLH-Zip) TF that belongs to the Myc supergene family and regulates gene expression by functioning as a homo- or heterodimeric TF (Steingrímsson et al., 2004). MITF is extensively involved in the regulation and signaling pathways associated with the development of melanocytes and retinal pigment epithelial cells (RPE) (Hou & Pavan, 2008). Mitf mutant mice display depigmented coat colors (reduced, spotty, or absent), and some of them might present with deafness and small or absent eyes because of impaired RPE morphogenesis (Hou & Pavan, 2008; Steingrímsson et al., 2004). The absence of intermediate cells and a decrease in cochlear potential in the stria vascularis were observed in mutant Mitf mice, thus possibly explaining the SNHI phenotype in patients with MITF variants (Huang et al., 2021). In humans, about 15% of WS2 patients possess MITF variants (Read & Newton, 1997). Some studies have described variable phenotypes and incomplete penetrance of each symptom in multiplex families (Alehabib et al., 2020; Lautenschlager et al., 1996), as was observed in the WSF-13 and WSF-14 families (Figure 2). The mild or absent manifestations of WS in individuals with MITF variants suggest that other modifying factors may affect the disease's expression (Lautenschlager et al., 1996). In our study, the novel missense MITF variant NM_198159.3:c.1052C>T caused an amino acid change in the third transactivation domain (AD3, Figure 1d) and was responsible for WS in the WSF-13 family.
In this study, we demonstrated the strength of NGS in identifying novel variants in WS-associated genes. Moreover, we also revealed that WSF-19 (GJB2), WSF-20 (SLC26A4, OMIM* 605646), and WSF-21 (OTOF, OMIM*603681) harbored variants associated with SNHI-related genes (Table S3), validating the use of NGS to clarify the underlying mechanism of isolated SNHI (Wu et al., 2013). However, this study had several limitations. First, we noted that three patients could not be genetically diagnosed with WS or other diseases. No candidate variant was detected in WSF-16, indicating that the causative variant may reside in regions other than the six WS-associated genes (Seco et al., 2015), the presence of structural variants (Jin et al., 2021; Wildhardt et al., 2013)or copy number variations (CNVs) (Bocángel et al., 2018; Wang et al., 2022; Wenzhi et al., 2015), or the existence of variants in non-coding regions (Bondurand et al., 2012), detection of which were beyond the limits of the NGS technique used in this study. Variants of uncertain significance were recognized in WSF-18 (MITF), suggesting that further functional study may be required to elucidate the exact effect of these variants. Second, we recruited a relatively small cohort size of 24 patients from a single population (Han Taiwanese), limiting the generalizability of the study results. Table S3 lists the phenotype and genotype information of six patients unable to be genetically diagnosed.
In conclusion, our results support that NGS is a useful procedure for diagnosing WS. With the help of NGS, we identified nine novel variants in SOX10, EDNRB, MITF, and PAX3 that may cause WS in Taiwanese patients. Furthermore, we validated the characteristics of incomplete penetrance and variable phenotypes in WS, suggesting that a more complex regulatory framework may govern the genetic expression of the disease.
ACKNOWLEDGMENTS
We thank all the patients and their relatives participating this study. The high-coverage targeted next-generation sequencing data was generated at the Labortary of Molecular Genetic Diagnostics in National Taiwan University Hospital. We also thank to National Center for High-performance Computing (NCHC) for providing computational and storage resources.
AUTHOR CONTRIBUTIONS
CYL designed and directed the study and drafted the manuscript; YMC performed the NGS experiment; MYL performed the Sanger sequencing; CYL, MYL, and YMC contributed equally to the data analyses; PHL, CJH, PLC, and CCW provided clinical resources and advices; PLC, CCW, and SJH critically reviewed the manuscript; SJH supervised the whole research.
FUNDING INFORMATION
This study was supported by research grants from the collaboration project of the National Taiwan University Hospital and National Taiwan University (UN110-040). This study was also funded by Health Promotion Administration, Ministry of Health and Welfare: 2022 Rare Disease Prevention Project.
CONFLICT OF INTEREST
The authors have no relevant financial or non-financial interests to disclose.
CONSENT TO PUBLISH
The authors affirm that human research participants provided informed consent for publication of the data related to their phenotypes and genotypes.
ETHIC STATEMENT
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Research Ethics Committees of the National Taiwan University Hospital (2021/11/09, No. 1103705165). All participants and/or their parents provided written informed consent.




