Characterization of CRB1 splicing in retinal organoids derived from a patient with adult-onset rod-cone dystrophy caused by the c.1892A>G and c.2548G>A variants
Abstract
Background
Mutations in the human crumbs homologue 1 (CRB1) gene are associated with a spectrum of inherited retinal diseases. However, functional studies demonstrating the impact of individual CRB1 mutations on gene expression are lacking for most variants. Here, we investigated the effect of two CRB1 variants on pre-mRNA splicing using neural retinal organoids (NRO) derived from a patient with recessive rod-cone dystrophy caused by compound heterozygous mutations in CRB1 (c.1892A>G and c.2548G>A).
Methods
The patient received ophthalmological examinations including multimodal imaging. NRO were differentiated from induced pluripotent stem cells (iPSCs) derived from the patient and a control subject. CRB1 transcripts were characterized by RT-PCR and Sanger sequencing.
Results
The Patient displayed retinal thickening with disorganization of retinal layers and preservation of para-arteriolar retinal pigment epithelium. Both patient and control iPSC produced NRO containing photoreceptor progenitor cells expressing CRB1 mRNA. Patient NRO expressed a novel CRB1 transcript displaying skipping of exon 6. CRB1 transcripts containing the c.2548G>A substitution in exon 7 were expressed in patient NRO.
Conclusions
Together, these results confirm the pathogenicity of the c.1892A>G and c.2548G>A CRB1 variants in a family with recessive adult-onset rod-cone dystrophy and further demonstrate the effects of these variants on pre-mRNA splicing. This data provide important insights into the pathogenic mechanisms associated with these variants.
1 INTRODUCTION
Mutations in the human crumbs homolog-1 (CRB1, OMIM#604210, NM_201253.2) gene are associated with a spectrum of inherited retinal diseases (IRD), including Leber congenital amaurosis 8 (LCA8), retinitis pigmentosa 12 (RP12), rod-cone dystrophy of varying age of onset, and foveal retinoschisis of adult onset (Bujakowska et al., 2012; den Hollander et al., 2001). These distinct phenotypes share the common features of photoreceptor and retinal pigment epithelium (RPE) degeneration with variable vascular abnormalities in the peripheral retina. Unique clinical features specifically found in CRB1-associated retinopathy are preservation of the para-arteriolar RPE (den Hollander et al., 2000), retinal telangiectasia with exudation (also referred to as Coats-like vasculopathy), as well as thickening of the retina and disorganized retinal layering (Jacobson et al., 2003).
The Genome Aggregation Database (gnomAD) currently lists 1833 known variants for the human CRB1 gene, including 910 missense substitutions, 317 synonymous substitutions, 109 3’/5’-UTR variants, 60 splice variants, 26 frameshifting mutations, 9 indels, 30 nonsense mutations, 371 intronic sequence variants and 1 start site mutation. Despite this significant progress in the identification of CRB1 gene variants, the pathogenicity of many of these variants remains unclear. A total of 323 disease-causing CRB1 variants are currently listed in the Human Genome Mutation Database (Stenson et al., 2017). In the absence of functional evidence, the clinical interpretation of these variants is somewhat influenced by in silico predictions according to American College of Medical Genetics and Genomics and Association for Molecular Pathology (ACMGG/AMP) guidelines (Richards, et al., 2015). While these in silico programs provide an excellent tool for predicting the clinical significance of novel gene variants, classifications obtained must be treated with caution until functional molecular evidence becomes available. With the ever-expanding catalogue of known variants in CRB1 and other disease-causing genes, it is essential that the effects of these mutations are established through molecular analyses. A clear understanding of the underlying mechanisms by which mutations exert their pathogenic effects can also inform genotype-phenotype correlations and guide future development of potential therapies.
Until recently, molecular characterization of IRD-causing mutations has been limited by a paucity of available retinal tissues from affected patients for analysis. Since many IRD genes are expressed specifically in the retina, access to patient retinal cells is essential for the characterization of IRD mutations at the RNA and protein level in the context of the affected cell type. The discovery of methods for the generation of induced pluripotent stem cells (iPSCs) from human somatic cells has provided the means for generating retinal cells from patients carrying disease-causing mutations in IRD genes (Chen et al., 2014). Recently, we reported the generation of iPSC lines from a patient with rod-cone dystrophy caused by compound heterozygous mutations (c.1892A>G and c.2548G>A) in CRB1 (Zhang et al., 2018). Interestingly, the c.1892A>G variant has been associated with a milder clinical phenotype (onset ≥10 years). In this study, we provide a detailed description of this patient's clinical phenotype and investigate the consequence of their mutations on transcript splicing using the patient's iPSC-derived retinal tissues.
2 METHODS
Both clinical and experimental aspects of this study were approved by the University of Western Australia (RA/4/1/7916, RA/4/20/5717) and the Sir Charles Gairdner Hospital (2001-053) Human Research Ethics Committees. Written consent was obtained from the patient and all procedures were carried out in accordance with the requirements of the National Health & Medical Research Council of Australia and the Declaration of Helsinki.
2.1 Clinical assessment
History and full ophthalmic examination including best-corrected visual acuity using the Early Treatment of Diabetic Retinopathy Study (ETDRS) letter chart and dilated fundus examination was undertaken. Multimodal retinal imaging included ultra-wide-field color fundus photography, green-light autofluorescence (AF) imaging (P200Tx and California, Optos plc, Dunfermline, UK), 30° and 55° scanning laser ophthalmoscopy including near-infrared reflectance (NIR), blue-light and near-infrared AF and spectral domain optical coherence tomography (SD-OCT, Spectralis OCT2 and Spectralis HRA2, Heidelberg Engineering, Heidelberg, Germany).
2.2 Genetic analysis and interpretation
Genetic analysis utilized genomic DNA extracted from peripheral blood samples collected and stored as detailed previously (Paterson et al., 2012). Analysis of proband genomic DNA targeted known variants in 28 genes associated with autosomal recessive retinitis pigmentosa (ARRP) using the Apex ARRP Array (version 5.3; performed by Asper Ophthalmics, Tartu, ESTONIA) coupled with haplotype analysis to exclude non-candidate genes (performed by the Australian Genome Research Facility (AGRF), Brisbane, Australia), as described previously (Paterson et al., 2012). Bidirectional Sanger sequencing of coding and flanking intronic regions of candidate genes CRB1, PDE6A, and RPE65 was performed on DNA of the proband and an affected sibling to detect novel candidate variants in these genes. The phase of CRB1 variants was established in parental DNA, and their presence in two siblings was ascertained by bidirectional Sanger sequencing (performed by ASPER Ophthalmics or AGRF). Variant nomenclature is in accordance with recommendations of the Human Genome Variation Society (den Dunnen et al., 2016unnen et al., 2016), with sequencing results aligned to the CRB1 reference sequence NM_201253.2 (OMIM#604210). Nucleotide 1 corresponds to the A of the ATG initiation codon.
Variant pathogenicity assessment encompassed in silico pathogenicity predictions, allele frequencies sourced from gnomAD (Karczewski et al., 2020), data from variation databases (LOVD/ClinVar/dbSNP) and the scientific literature, and the clinical diagnosis. In silico pathogenicity assessment utilized Mutation Taster (Schwarz, Rodelsperger, Schuelke, & Seelow, 2010), SIFT (Ng & Henikoff, 2001), PolyPhen2 (Adzhubei et al., 2010), Align GVGD (Tavtigian, Byrnes, Goldgar, & Thomas, 2008), REVEL (Ioannidis et al., 2016), M-CAP (Jagadeesh et al., 2016) and the Alamut Visual splicing prediction module (Version 2.11; Interactive Biosoftware). Pathogenicity interpretation was performed in accordance with ACMGG/AMP guidelines (Richards et al., 2015). Additionally, in silico splicing analyses were performed using Human Splicing Finder, Version 3.1 (Desmet et al., 2009) and EX-SKIP (Raponi et al., 2011).
2.3 Cell culture and differentiation
Patient iPSC (LEIi006-A) and the normal control iPSC line (Cat#A18945, Thermo Fisher Scientific) were cultured in feeder-free conditions, on geltrex (Thermo Fisher) coated culture plates in mTeSR1 medium (StemCell Technologies), as previously described (Zhang et al., 2018). The mutations found in peripheral blood DNA were also confirmed in these patient-derived iPSC lines. For directed differentiation into neural retinal organoids (NRO) we followed a previously published protocol (Mellough et al., 2015, 2019). Patient and control iPSC colonies were dissociated into small pieces by EDTA buffer (0.5 mM EDTA and 30 mM NaCl in DPBS (-Ca++, -Mg++) and then cultured in the DMEM/F-12 medium (Thermo Fisher) with decreasing concentrations of KnockOut Serum Replacement (KOSR, Thermo Fisher) (20% for the first 5 days, 15% until day 12, 10% until day 35) containing MEM Non-Essential Amino Acids Solution (NEAA; Thermo Fisher), B27 (Thermo Fisher) and IGF-1 (StemCell).
2.4 Transcript analysis
Total mRNA was isolated from early (day 35) NRO using TRIZOL and cDNA was synthesized using the RT2 First Strand Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Partial cDNA sequences were amplified with Q5 High-Fidelity Polymerase (New England Biolabs, Ipswich, Massachusetts, USA) using primers targeting exons 3–7 or exons 7–8. Primer sequences are listed in Table S1. PCR products were resolved on a 2% agarose gel. The 50 bp DNA Ladder (New England BioLabs) was used as a molecular size marker. Gels were imaged using the ChemiDoc XRS+imaging system (BioRad). After purification with the Gel Purification and PCR Clean-up System (Promega), PCR fragments were assessed by Sanger sequencing (AGRF).
2.5 Immunostaining analysis
Retinal organoids were fixed with 4% paraformaldehyde, washed, then permeabilized using phosphate-buffered saline (PBS) with 0.1% Triton X-100 for 15 min. The samples were then incubated in 5% BSA in PBS for 1 hr at room temperature. Primary antibodies were applied at 4°C overnight. Secondary antibodies were applied for 2 hours at room temperature. Nuclei were stained with DAPI. The following primary antibodies were used in this study: Mouse anti-Pax6 (1:100, Abcam, AB78545), Rabbit anti-Recoverin (1:200, Merck, AB5585). Secondary antibodies used in this study included Alexa Fluor 546 Goat anti-mouse IgG (1:500, Molecular Probes, A-11003), and Alexa Fluor 546 Goat anti-rabbit IgG (1:500, Molecular Probes, A-11035).
2.6 Transmission electron microscopy (TEM) analysis
Retinal organoids were fixed in 4% paraformaldehyde, 2% glutaraldehyde in 0.1 M phosphate buffer, pH 7.4. After 24 hours, they were post-fixed in 1% osmium tetroxide in the same buffer for 6 minutes (2 minutes on, 2 minutes off and 2 minutes on) with biowave (PELCO). Samples were then dehydrated in a graded series of ethanol and acetone followed by embedding in resin containing Procure 812, Araldite 502, DDSA and BDMA. Ultrathin (100 nm) sections were prepared and examined with a JEM2100 electron microscope (JEOL).
2.7 Quantitative PCR
Total mRNA was isolated from NRO cultures using TRIZOL and cDNA was synthesized using the RT2 First Strand Kit (Qiagen). Quantitative PCR (qPCR) was performed using RT2 SYBR Green qPCR Mastermix (Qiagen) and the CFX Connect Real-Time System (BioRad). Data were analyzed using the ΔΔCT method. Gene expression values were normalized to expression levels measured in LEIi005-A iPSC. Sequences for primers used in this study are listed in Table S1.
3 RESULTS
3.1 Clinical presentation
The patient was a 50-year-old female initially diagnosed with rod-cone dystrophy and macular cysts at the age of 21. Her presenting Snellen visual acuity was 20/30 in both eyes. Fundus examination in the early stages of the disease showed bilateral cystoid maculopathy with numerous white dots in the periphery resembling fundus albipunctatus. Over the 30 years of follow-up, her vision declined to perception of light in both eyes and the majority of the retina was replaced by heavy intraretinal pigmentation sparing the para-arteriolar regions in the periphery (Figure 1a-b). There was gross thickening of the retina due to disorganized retinal layering (Figure 1c). She also developed non-visually significant posterior subcapsular cataract at age 46.
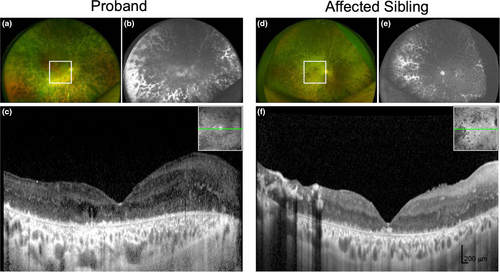
The proband's brother presented at age 18 with a visual acuity of 20/40 in both eyes and peripheral retinal schisis that led to an initial clinical diagnosis of Goldman-Favre syndrome. He had bilateral cataract extraction at the age of 36, and 9 years later, his vision was reduced to perception of light. He also had heavy intraretinal pigmentation with para-arteriolar sparing (Figure 1d-e) and thickened retina (Figure 1f). Neither the proband nor the affected sibling manifested variable vascular abnormalities or retinal telangiectasia with exudation. The proband's parents underwent similar clinical examinations, with no retinal abnormalities detected (data not shown).
3.2 Genetic analysis and pathogenicity assessment
Proband DNA was analyzed by Apex ARRP Array, coupled with haplotype analysis to further identify candidate genes. Subsequent Sanger sequencing of PDE6A and RPE65 excluded novel candidate variants in these genes. Two variants in CRB1 were identified: c.1892A>G and c.2548G>A. Segregation analysis confirmed the paternal (former) and maternal (latter) origin of these variants and their segregation with disease (Figure 2a). These missense variants had predicted amino acid substitutions p.(Tyr631Cys) and p.(Gly850Ser), respectively, expected to affect conserved laminin A/G-like domains (Figure 2b).
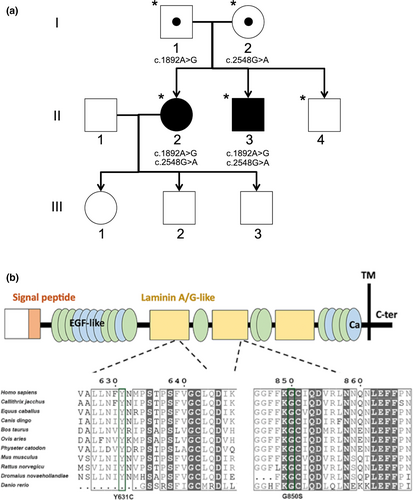
In silico pathogenicity analysis provided conflicting results for c.1892A>G: despite aberrant splicing predictions, Mutation Taster returned a benign prediction, as did SIFT, PolyPhen 2 and REVEL. AlignGVGD and M-CAP returned a possibly pathogenic result, and EX-SKIP predicted the mutant allele was more likely to cause exon skipping than the wild-type allele. Analysis of this variant using Human Splicing Finder (HSF, version 3.1) predicted potential alteration of splicing due to the introduction of an exonic splice silencer (ESS) site and a cryptic donor splice site (DSS) in exon 6 (Figure S1). Comparison of normal and c.1892A>G CRB1 splice site sequences in exon 6 revealed the cryptic DSS present in the mutant sequence had a lower HSF score (67.55) than the natural DSS (94.75), suggesting it may not be a strong competitor for the natural DSS. The strength of the Fas-ESS hexamer sequence generated by the c.1892A>G variant could not be assessed by the HSF software, but may promote skipping of exon 6 during pre-mRNA splicing. Interrogation of variation databases and the literature indicated that this variant is rare (allele frequency = 0.000008), and is reported as causative for autosomal recessive retinal dystrophy or retinitis pigmentosa (Hettinga, van Genderen, Wieringa, Ossewaarde-van Norel, & de Boer, 2016; Talib et al., 2017). Recent reports have additionally provided in vitro evidence of mislocalized CRB1 protein and misplaced photoreceptors in patient-derived retinal organoids subsequent to this variant occurring in trans with a nonsense CRB1 variant (Quinn et al., 2019). The scientific literature, thus, suggests a pathogenic status for this variant.
In contrast, the c.2548G>A variant was predicted to be pathogenic by all in silico algorithms utilized. Despite some predictions of altered splicing motifs, EX-SKIP predicted the mutant allele was unlikely to induce exon skipping. HSF predicted potential alterations of splicing due to the disruption of an SF2/ASF exonic splicing enhancer (ESE) motif in exon 7 (Figure S2). However, the impact of the predicted loss of this ESE element is unclear. This rare variant (allele frequency = 0.000025) has been reported as causative for ARRP with or without preservation of para-arteriolar RPE, and LCA8 (Clark et al., 2010; den Hollander et al., 2004; Henderson et al., 2011). Taken together, we considered both of these variants likely candidates for disease (Table S2), although the effects of these variants at the mRNA or protein level had not been demonstrated.
3.3 Differentiation of photoreceptor progenitor cells from patient and control iPSCs
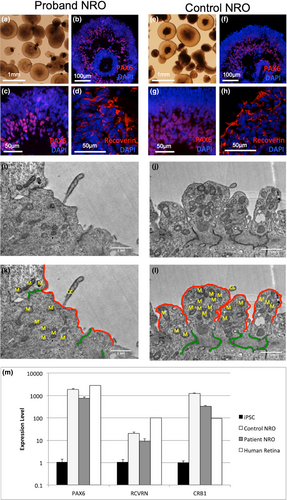
Given the in silico predictions of pathogenicity, we hypothesized that one or both variants might lead to alternative splicing of the CRB1 gene, leading to a deficit in functional CRB1 protein expression. To test this hypothesis, we generated neural retinal organoids (NRO) from control iPSCs and the LEIi006-A iPSC line previously generated from proband dermal fibroblasts (Zhang et al., 2018). After 35 days of differentiation, both control (Figure 3a-d) and patient (Figure 3e-h) iPSCs produced laminated NRO resembling early optic cups. Immunostaining demonstrated the localization of retinal ganglion cells (RGCs) and amacrine cells expressing PAX6 in the center of the organoid surrounded by a dense layer of neural retinal progenitor cells (Figure 3b-c and f-g). Clusters of immature photoreceptor progenitor cells (PPCs) expressing recoverin were identified in the developing retinal progenitor cell layer (Figure 3d and h). Ultrastructural examination of NRO by transmission electron microscopy identified photoreceptor inner segments (IS) on the surface of both control (Figure 3i,k) and patient (Figure 3j,l) NRO. Junctional complexes were observed in the subapical region of PPCs from both genotypes (Figure 3i-l). Expression of PAX6, RCVRN, and CRB1 in NRO was confirmed by qPCR analysis of day 35 NRO cDNA, using adult human retinal cDNA as a positive control (Figure 3m). All three retinal markers were upregulated in NRO and human retina compared with undifferentiated iPSC. PAX6 was expressed at similar levels in NRO and adult human retina, while RCVRN expression was lower in NRO than in adult human retina, consistent with the low numbers of PPCs observed at this early time point. CRB1 mRNA expression was increased in NRO compared with human retina. Patient NRO expressed moderately lower levels of PAX6, RCVRN, and CRB1 than control NRO, likely reflecting a lower efficiency of NRO induction from the patient iPSC line. Together, these results demonstrate the successful generation of early-stage NRO containing an inner RGC/amacrine cell layer and an outer retinal progenitor cell layer containing clusters of immature PPCs.
3.4 The CRB1 c.1892A>G variant leads to exon skipping
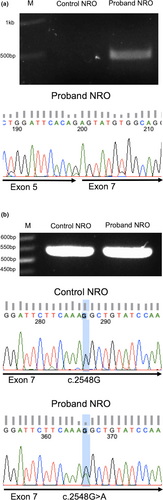
To examine splicing of the CRB1 gene in patient and control PPCs, we amplified partial cDNA sequences from early NRO using primers targeting exons 3-7. We detected a 0.5 kb band in patient, but not control, NRO that corresponded to a novel CRB1 transcript lacking exon 6 (Figure 4a). Sanger sequencing of this fragment demonstrated the splicing of exon 5 to exon 7, producing an in-frame deletion in the CRB1 coding sequence. If translated, the transcript would produce a protein with a 319-amino acid internal deletion, p.(Gly391_Arg709del). Since full-length CRB1 cDNA sequence was not detected in this experiment due to the large size of the full-length product (1.7 kb), we performed additional PCR screening using primers targeting exon 6 and the exon 6-7 junction to determine whether CRB1 transcripts containing exon 6 were produced from the paternal c.1892A>G allele. A single band amplified from proband NRO displayed a single “A” peak at position c.1892, indicating the product was derived from mRNA transcribed from the maternal c.2548G>A allele (Figure S3). These results indicate that the c.1892A>G variant results in the genesis of an ESS, which leads to exon 6 skipping during pre-mRNA splicing. Based on these results, we propose a new ACMGG/AMP classification (Pathogenic (ia)) for this variant (Table S2).
3.5 The CRB1 c.2548G>A variant is a missense mutation
Using primers targeting exons 7-8, we detected CRB1 transcripts in both patient and control NRO. Purification and sequencing of the patient NRO PCR product revealed a heterozygous c.2548G>A signal demonstrating the presence of CRB1 mRNA transcribed from both alleles (Figure 4b). These results indicate that CRB1 mRNA transcripts carrying the c.2548G>A missense mutation are produced in patient retinal cells. If translated, these transcripts are predicted to produce mutant CRB1 proteins carrying the p.Gly850Ser amino acid substitution.
4 DISCUSSION
In this study, we used iPSC-derived NRO to demonstrate altered CRB1 splicing in the retinal cells of a patient with the compound heterozygous CRB1 mutations c.[1892A>G];[2548G>A]. The c.1892A>G variant, previously classified as a missense variant, was shown to cause exon 6 skipping, whereas the c.2548G>A variant produced CRB1 transcripts encoding the p.Gly850Ser missense mutation. These results are supported by the clinical observation of impaired retinal maturation in OCT scans from both the proband and her affected sibling. These rare CRB1 variants have previously been associated with severe retinal dystrophies, and are present in variation databases with clinical assertions of VUS or pathogenic. The conflicting in silico results for the c.1892A>G variant likely reflect the perspective of these programs, with those assessing nucleotide and amino acid substitutions considering this variant to be a neutral change. Mutation Taster, which also assesses splicing changes, utilizes NNSplice, which analyses the structure of donor and acceptor splice sites and not changes to exonic splice enhancers/silencers, explaining the benign prediction. In contrast, EX-SKIP and HSF, which take into account the ESE/ESS profile, predicted the possibility of exon skipping.
In the literature, the CRB1 c.1892A>G variant has been associated with retinal dystrophy and ARRP (Hettinga et al., 2016; Quinn et al., 2019; Talib et al., 2017), while CRB1 c.2548G>A has been associated with LCA8, and ARRP with or without preserved para-arteriolar RPE (Clark et al., 2010; den Hollander et al., 2004; Henderson et al., 2011). In a recent study, Quinn et al. characterized human fetal retinal tissues and iPSC-derived NRO and showed that expression of crumbs complex proteins in vivo was closely recapitulated by the in vitro human NRO model (Quinn et al., 2019). While CRB2 was abundantly detected in the subapical region of PPCs and Muller glia of first trimester and early (2 month) NRO, CRB1 immunoreactivity was sporadic and not localized to the subapical regions at these time points. CRB1 immunoreactivity and subapical localization increased with retinal maturation. Additionally, these authors characterized NRO derived from three patients with CRB1 mutations, one of whom carried the c.1892A>G mutation. Late stage (6 month) NRO from all three patient iPSC lines displayed features previously associated with crumbs complex deficiencies in mouse models, including disrupted outer limiting membrane formation and mislocalization of photoreceptors (Quinn et al., 2019).
In the present study, we did not observe overt defects in early NRO, consistent with the limited CRB1 protein expression in early retinal development. However, since early NRO expressed CRB1 mRNA, we were able to determine the effects of two mutations on splicing of the CRB1 transcript. We identified putative protein-coding transcripts expressed from both alleles (Figure 4).
The allele carrying the c.2548G>A variant produced a CRB1 transcript predicted to encode a mutant CRB1 protein incorporating a single amino acid substitution, p.(Gly850Ser). This glycine residue was previously shown to be conserved across all nine laminin A/G domains found in human CRB1, mouse Crb1 and Drosophila Crb (den Hollander et al., 2004), suggesting a critical role in protein structure and function. Replacement of the small, uncharged glycine residue with the larger, polar serine may disrupt protein folding or interfere with the protein–protein interactions mediated by this domain.
Consistent with HSF and EX-SKIP predictions, which showed the generation of an ESS in exon 6 by the c.1892A>G variant, we also demonstrated the presence of exon 6-skipped CRB1 transcripts in patient NRO harboring this mutation. Removal of exon 6 from the CRB1 transcript results in an in-frame deletion of 957 bp of sequence encoding 319 amino acids that include a calcium-binding EGF-like domain, two EGF-like domains and a Laminin A/G domain (den Hollander et al., 2004). These domains play an essential role in protein–protein interactions and calcium binding (Selander-Sunnerhagen et al., 1992), suggesting the truncated protein might have reduced function. However, the presence of intact transmembrane and C-terminal domains in the mutant protein may preserve some ability to interact with other core crumbs complex proteins. Indeed, the late age of onset in the proband supports the presence of residual CRB1 function in the retina: in contrast to patients with null alleles who manifest blindness in the first year of life, our patients maintained visual function for at least the first decade.
The previous association of the c.2548G>A variant with early onset LCA suggests the missense substitution likely has a severe effect on CRB1 protein function. To date, the c.1892A>G variant has only been reported in patients with disease onset in the second to fifth decade of life, suggesting it could have a more moderate effect. Moving forward, characterization of expression, localization, and interactions of these putative mutant CRB1 proteins in late-stage NRO is required to ascertain the level of residual CRB1 function from the in-frame deletion of exon 6 and the p.(Gly850Ser) substitution.
In conclusion, our study confirmed the pathogenicity of the c.1892A>G and c.2548G>A variants in CRB1 in a family with recessive adult-onset rod-cone dystrophy. We demonstrated the utility of patient-NRO in confirming the effects of c.1892A>G and c.2548G>A variants on transcript splicing. Future transcript analysis in CRB1 patient-NRO may allow characterization of more exonic and intronic mutations that lead to altered splicing, thus revealing a molecular basis to explain the clinical phenotype and an opportunity for development of personalized treatment using splicing interventions.
ACKNOWLEDGMENTS
This work was funded by the Australian National Health and Medical Research Council (GNT1188694, MRF1142962, GNT1116360) and Ophthalmic Research Institute of Australia. The authors acknowledge the generous donations from the Miocevich family, Saleeba family, McCusker family, the Australian Foundation for the Prevention of Blindness and the University of Western Australia (International Postgraduate Research Scholarship). The Australian Inherited Retinal Disease Registry acknowledges the financial support of Retina Australia.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHOR CONTRIBUTION
SM, FKC, and XZ contributed to project design and wrote the manuscript draft. All authors contributed to data collection and analysis as well as revision and final approval of the published manuscript.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




