Resistance to abemaciclib is associated with increased metastatic potential and lysosomal protein deregulation in breast cancer cells
Abstract
Cyclin dependent kinase 4 and 6 inhibitors such as abemaciclib are routinely used to treat metastatic estrogen receptor positive (ER+) breast cancer. However, adaptive mechanisms inhibit their effectiveness and allow for disease progression. Using ER+ breast cancer cell models, we show that acquired resistance to abemaciclib is accompanied by increase in metastatic potential. Mass spectrometry-based proteomics from abemaciclib sensitive and resistant cells showed that lysosomal proteins including CTSD (cathepsin D), cathepsin A and CD68 were significantly increased in resistant cells. Combination of abemaciclib and a lysosomal destabilizer, such as hydroxychloroquine (HCQ) or bafilomycin A1, resensitized resistant cells to abemaciclib. Also, combination of abemaciclib and HCQ decreased migration and invasive potential and increased lysosomal membrane permeability in resistant cells. Prosurvival B cell lymphoma 2 (BCL2) protein levels were elevated in resistant cells, and a triple treatment with abemaciclib, HCQ, and BCL2 inhibitor, venetoclax, significantly inhibited cell growth compared to treatment with abemaciclib and HCQ. Furthermore, resistant cells showed increased levels of Transcription Factor EB (TFEB), a master regulator of lysosomal-autophagy genes, and siRNA mediated knockdown of TFEB decreased invasion in resistant cells. TFEB was found to be mutated in a subset of invasive human breast cancer samples, and overall survival analysis in ER+, lymph node-positive breast cancer showed that increased TFEB expression correlated with decreased survival. Collectively, we show that acquired resistance to abemaciclib leads to increased metastatic potential and increased levels of protumorigenic lysosomal proteins. Therefore, the lysosomal pathway could be a therapeutic target in advanced ER+ breast cancer.
Abbreviations
-
- ABBY
-
- Pilot Trial of ABemacicliB or Abemaciclib and HydroxYchloroquine to Target Minimal Residual Disease in Breast Cancer Patients
-
- Baf
-
- Bafilomycin
-
- BCL2
-
- B cell lymphoma 2
-
- CCS
-
- charcoal stripped calf serum
-
- CD68
-
- lysosomal/endosomal-associated membrane glycoprotein LAMP4
-
- CDK4
-
- cyclin dependent kinase 4
-
- CDK4/6i
-
- cyclin dependent kinase 4/6 inhibitors
-
- CDK6
-
- cyclin dependent kinase 6
-
- CLEAR
-
- coordinated lysosomal expression and regulation
-
- CTSA
-
- cathepsin A
-
- CTSD
-
- cathepsin D
-
- CTSZ
-
- cathepsin Z
-
- DMEM
-
- Dulbecco's Modified Eagle Medium
-
- DMSO
-
- dimethyl sulfoxide
-
- DTT
-
- dithiothreitol, 1,4-dithiothreitol
-
- ECM
-
- extracellular matrix
-
- ER
-
- estrogen receptor
-
- EtOH
-
- ethanol
-
- FBS
-
- fetal bovine serum
-
- HCQ
-
- hydroxychloroquine
-
- HER2
-
- human epidermal growth factor receptor 2
-
- HR
-
- hormone receptor
-
- IMEM
-
- Improved Minimum Essential Medium
-
- LAMP1
-
- lysosomal associated membrane protein 1
-
- LAMP4
-
- lysosomal associated membrane protein 4
-
- LC3A
-
- MAP1LC3A
-
- LC3B
-
- MAP1LC3B
-
- LDCD
-
- lysosome dependent cell death
-
- LMP
-
- lysosomal membrane permeabilization
-
- MITF
-
- microphthalmia
-
- OBT
-
- obatoclax
-
- p62
-
- SQSTM1
-
- PALVEN
-
- A phase 1b study of palbociclib, letrozole and venetoclax in estrogen receptor, BCL2-positive metastatic breast cancer
-
- PARP
-
- poly(ADP-ribosyl)transferase
-
- PBS
-
- phosphate-buffered saline
-
- RB1
-
- retinoblastoma 1
-
- SDS
-
- sodium dodecyl sulfate
-
- TBST
-
- tris-buffered saline in Tween 20
-
- TFE3
-
- Transcription Factor Binding to IGHM Enhancer 3
-
- TFEB
-
- Transcription Factor EB
-
- TFEC
-
- Transcription Factor EC
-
- TNBC
-
- triple negative breast cancer
-
- VNX
-
- venetoclax
1 INTRODUCTION
Resistance to Cyclin dependent kinase 4/6 inhibitors (CDK4/6i) is a current clinical problem in treating hormone receptor positive (HR+)/HER2− breast cancer. Three CDK4/6i are currently FDA approved: palbociclib, ribociclib, and abemaciclib. These drugs inhibit cell cycle dependent kinases, CDK4 and CDK6, from binding with Cyclin D1 to prevent RB1 phosphorylation and induce G1 arrest. It is estimated that 20% of patients with tumors resistant to endocrine therapy have intrinsic resistance to CDK4/6i, and all patients eventually go on to acquire resistance to these drugs over time.1, 2 Currently, there is no drug specifically intended for use in CDK4/6i resistant breast cancer. Abemaciclib (LY2835219; Verzenio) is the only CDK4/6i currently approved as a monotherapy in metastatic breast cancer,3 and has been recently been approved for early high-risk breast cancer in combination with endocrine therapy.4 Abemaciclib is 5-times more potent against CDK4 than palbociclib or ribociclib5 and can cross the blood brain barrier.6 Its relatively weak activity against CDK6 adds to its clinical value, as patients do not experience the CDK6 specific-side effect of neutropenia, and thus, no drug holiday is needed.
In the nucleus, CDK4/6 phosphorylates Transcription Factor EB (TFEB) and Transcription Factor Binding to IGHM Enhancer 3 (TFE3), master regulators of lysosomal-autophagy pathway genes. Phosphorylation of TFEB/TFE3 causes them to be shuttled to the cytoplasm, and thus TFEB/TFE3-mediated gene transcription of the Coordinated Lysosomal Expression and Regulation (CLEAR) gene network is prevented.7, 8 TFEB/TFE3 and the CLEAR network tightly regulate the activity of many enzymes to control lysosomal functions such as exocytosis and autophagy. Abemaciclib mediated inhibition of CDK4/6 can prevent nuclear phosphorylation of TFEB/TFE3 and increase lysosomal biomass and function, which mediates a novel lysosomal-mediated form of cell death in sensitive cells.7, 9 In a subset of triple negative breast cancer (TNBC) cells with intact RB1, TFEB mediated transcription of lysosomal genes was shown to increase lysosomal mass and enabled these TNBC cells to intrinsically resist palbociclib by sequestering the drugs into the lysosomes.10 However, the mechanism by which estrogen receptor positive (ER+) breast cancer cells use lysosomes to acquire resistance to long-term CDK4/6i treatment remains unknown.
The relationship between CDK4/6 inhibition and lysosomal mass/function is particularly interesting in the context of advanced disease. Though drug resistance is often seen in the advanced setting, only recently has research started to investigate connections between drug resistance and cancer metastasis.11, 12 Lysosomes have been implicated in metastasis in multiple ways. The surface of highly metastatic cancer cells has increased expression of lysosomal-associated membrane proteins.13 Likewise, cathepsins, proteolytic lysosomal enzymes, can degrade the extracellular matrix to facilitate invasion and thus have been associated with metastasis.14-17 Given this evidence and the recent line of thought that drug resistance is directly linked to metastasis, we hypothesized that deregulated activity in abemaciclib resistant cells causes cells to gain more migratory/invasive abilities. In this study, we used ER+ murine and human breast cancer cells with acquired resistance to abemaciclib to show that resistant cells with altered lysosomal proteins have increased metastatic behavior in vitro. As a result, combination of a lysosome-targeting drug and abemaciclib sensitizes resistant cells by inducing lysosomal membrane permeabilization (LMP).
2 MATERIALS AND METHODS
2.1 Reagents
Abemaciclib (LY2835219), hydroxychloroquine (HCQ) (NSC 4375), and bafilomycin (Baf) A1 (NSC 381866) were purchased from Cayman Chemical Company. Palbociclib (PD0332991) and ribociclib (LEE011) were purchased from Selleck Chemicals. All other reagents were purchased from Sigma Aldrich. Abemaciclib was dissolved in ethanol (EtOH). All other drugs were dissolved in dimethyl sulfoxide (DMSO). For in vitro assays, the negative control was 0.02% EtOH or DMSO.
2.2 Cell culture
Murine C57BL/6 PyMT-B6 and PyMT-Bo1 ER+ breast cancer cell lines were established as described previously.18 PyMT-Bo1 cells were derived from bone metastasis of PyMT-B6 tumors in vivo. MCF7 and LCC1 cells were obtained from Georgetown University Medical Center Tissue Culture Shared Resource. LCC1 cells were originally established from MCF7 cells and are estrogen-independent.19 Abemaciclib, palbociclib, and ribociclib-resistant cell lines were generated from PyMT-B6, PyMT-Bo1, and LCC1 cells in respective complete media supplemented with 1μM abemaciclib, palbociclib, or ribociclib for 6+ months. PyMT cells were cultured in Dulbecco's Modified Eagle Medium (DMEM; Thermo Fisher Scientific) and supplemented with 10% fetal bovine serum (FBS; Life Technologies) and penicillin-streptomycin (Thermo Fisher Scientific). MCF7 cells were cultured in Improved Minimum Essential Medium (IMEM; Thermo Fisher Scientific) supplemented with 10% charcoal-stripped calf serum (CCS; Life Technologies) and 10% estradiol.20 LCC1 cells were cultured in IMEM supplemented with 5% CCS. All cell lines were maintained in a humidified atmosphere with 5% CO2 at 37°C. All cells were authenticated by DNA fingerprinting and regularly tested for Mycoplasma infection.
2.3 Cell proliferation assay
For murine PyMT cell proliferation studies, cells were seeded with 1000 cells per well in 96-well plastic tissue culture plates per cell line. At 24 h postplating, cells were dosed with 0.02% vehicle (EtOH or DMSO) or various concentrations of the indicated drugs for 72 h. For human LCC1 cell proliferation studies, cells were seeded with 3500−10,000 cells per well and cells were treated with vehicle or indicated drugs for 6 days. For crystal violet staining, plates were rinsed once with 1x PBS to remove cellular debris. Then, 100 μL crystal violet was added to each well and rocked at room temperature for 1 h. To remove excess stain, each plate was rinsed 5−10x with H2O. The plates were left at room temperature to dry overnight, then were rehydrated with 100 μL 0.1 M sodium citrate buffer in 50% EtOH. Plates were measured using a Vmax kinetic microplate reader (Molecular Devices Corp.) with an absorbance of 560 nm. Results were normalized to vehicle. All experiments were repeated at least three times with n = 5−10 technical replicates for each condition.
2.4 Western blot analysis
Cells were washed once with cold 1x PBS. They were then lysed in radio-immunoprecipitation assay buffer supplemented with PhosSTOP phosphatase and CompleteMini protease inhibitors (Roche, Switzerland) for protein extraction. Proteins were separated by polyacrylamide gel electrophoresis using 4−12% gradient gels followed by protein transfer onto nitrocellulose membranes with iBLOT2 (Thermo Fisher Scientific). Membranes were blocked in 5% nonfat dry milk made in Tris-buffered saline with Tween-20 (TBST) and rocked at 4°C with the respective primary antibodies overnight. Secondary antibodies conjugated with horseradish peroxidase and SuperSignal West Dura Extended Duration Substrate (Thermo Fisher Scientific) were used to detect proteins of interest. The following antibodies were purchased from Cell Signaling Technology: CDK4 (#12790), CDK6 (#3136), phospho-RB1 (S780) (#3590), and Cyclin D1 (#2978), LC3B (#2775), Cleaved PARP (#9548 S), B cell lymphoma 2 (BCL2) (#3498 T), TFEB (#83010 S), and cathepsin D (CTSD) for human cell lysates (#2284). ER alpha (#ab108398) and RB1 (#ab181616) antibodies were purchased from Abcam. P62 (#610832) was purchased from BD Biosciences. β-tubulin (#T7816) was acquired from Sigma-Aldrich. Actin (#sc-47778) antibody was purchased from Santa Cruz Biotech. Cathepsin A (CTSA) for human cell lysates (#15020-1-AP) was purchased from Proteintech. CTSD for mouse cell lysates (#MAB1123-SP) and CTSA for mouse cell lysates (AF1029-SP) were bought from R&D Systems. Secondary antibodies for mouse (#7076 S) and rabbit (#7074 S) were purchased from Cell Signaling. Goat (#R-21459) and rat (#31470) secondary antibody was purchased from Thermo Fisher Scientific.
2.5 Migration and invasion assays
For migration assays, 1 × 104 cells in 100 μL media were plated in the top of the Corning filter membrane insert (VWR) sitting inside of a 24 well tissue culture plate. For invasion assays, 0.5 mL warm serum free media (SFM) was added on top of the Corning GFR Matrigel inserts (VWR). Matrigel was allowed to rehydrate for 2 h at 37°C. Then, the media was carefully removed so as to not disturb the matrigel. 1 × 104 cells in 100 μL media were plated in the top of the matrigel insert. Both assays used the same protocol after this point. Plates were incubated at 37°C for 10 min. Next, complete media was added into the lower chamber of the well. For invasion assays with TFEB siRNA or drug, the protocol remained the same except both the cells in SFM and the complete media at the bottom of the well contained 20 nM TFEB siRNA or the indicated drug. For assays with murine cells, plates were incubated for 24 h at 37°C. For assays with human cells, plates were incubated for 72 h at 37°C. Inserts were removed, and excess cells that did not move through the membrane/matrigel were gently removed from the surface using a cotton-tipped applicator. A Differential Quik III Stain Kit (Electron Microscopy Sciences) was used to stain the cells trapped in the membrane or matrigel. Migration or invasion were quantified by counting four fields at a magnification of 10x using a bright field microscope. Each experiment was repeated at least three times.
2.6 Mass spectrometry-based proteomic analysis
PyMT-B6, PyMT-B6-abemaR, PyMT-Bo1, and PyMT-Bo1-abemaR cells were plated and grown in 10 cm2 dishes in complete growth medium to 70% confluence. MCF7 cells were plated in 10 cm2 dishes for 24 h and treated with vehicle (0.02% EtOH) alone or 1 μM abemaciclib for 72 h. Four biological replicates were used for all cell lines. For proteomics analysis, cells were washed 3x with cold PBS, scraped down, centrifuged at 4°C followed by a cold PBS wash for another 3 times. Equal amount of proteins from each sample were processed for proteomics, with a procedure published previously.21 In brief, samples resuspended in 5% SDS buffer were reduced with DTT and alkylated, followed by digestion on a S-Trap column (ProtiFi, LLC) with sequencing-grade Lys-C/trypsin (Promega). The resulting peptides were analyzed with a nanoAcquity UPLC system (Waters) coupled with Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher), with a procedure published previously.21 All data were acquired with data independent acquisition (DIA) mode. DIA data files were processed as described previously.20 In brief, the Spectronaut software version 15 (Biognosys) and a hybrid library were used (with default settings). Proteins identified and quantified were exported for comparison. Unpaired two-tailed Student's t-tests were used to define significant differentially expressed proteins (with fold changes > 1.5 or <0.67; Q values < 0.05).
2.7 Growth of primary tumors in vivo
PyMT-B6, PyMT-B6-abemaR, PyMT-Bo1 or PyMT-Bo1-abemaR cells (1 × 105) were suspended in Matrigel (BD Biosciences) and orthotopically injected into the mammary fat pad of 8-week-old female C57BL/6 mice (n = 5) (Charles River). Body weight and tumor size were measured twice a week for 2 weeks, at which point, primary tumors were excised and processed for histology. Hematoxylin and eosin (H&E) stained slides were evaluated by a veterinary pathologist who evaluated clusters of degenrating or dead cells within tumors to determine tissue necrosis at our Histopathology and Tissue Shared Resource.
2.8 Transfection with TFEB siRNA
Cells were plated in six-well (for protein assessment) or 96-well (for proliferation assay) plates. TFEB siRNAs (siGENOME, M-050607-00 and ON-TARGETplus, L-050607-02, mixture of four siRNA), or scrambled negative control were purchased from Dharmacon, Inc. siRNAs were transfected into the cells using RNA iMAX (ThermoFisher Scientific). Any indicated treatments were added 24 h post-transfection. Cells were lysed or plates were ended 72 h post-transfection. siRNA targeted the following sequences of TFEB, siGENOME: “CUACGAAGAUGAUGAAUAC” “CAAGUUUGCUGCCCACGUG” “GCAGAUGCCUAACACGCUG” “ACUUAAUGCUCCUAGAUGA”; ON-TARGETplus: “GGAUCAAGGAGCUGGGAAU,” “CAUCAGAAGGUUCGGGAGU,” “GGUGUGAAGUAGCCGCCUA,” “CUAAUUGAGAGAAGACGCA.”
2.9 Cell cycle and cell death assays
2.9.1 Cell cycle
Cells were plated at 1 × 105 in 10 cm2 dishes. Cells for time = 0 h were grown in complete media and collected the following day. Treatment cells were grown in complete growth medium for 24 h before being treated with the indicated drug. Cells were then fixed in EtOH and analyzed by the Flow Cytometry Shared Resource according to the method of Vindelov et al.22
2.9.2 Apoptosis
For the apoptosis assay, 1 × 105 cells were plated in 10 cm2 dishes and treated with vehicle, 1 μM abemaciclib, 5 μM HCQ, or the combination for 72 h. Cells were stained with Annexin V-fluorescein isothiocyanate and propidium iodide, respectively (Thermofisher Scientific) according to the manufacturer's protocol and fluorescence was measured by the Flow Cytometry Shared Resource at Georgetown University Medical Center. Each experiment was repeated at least three times.
2.9.3 Autophagy
For the autophagy assay, 5 × 104 cells were plated in 12 well plates and treated with vehicle, 1 μM abemaciclib, 5 μM HCQ, or the combination for 72 h. Starvation control cells were grown in SFM for 72 h. Cells were stained with CYTO-ID green detection reagent (Enzo Life Sciences) according to the manufacturer's protocol and fluorescence was measured by the Flow Cytometry Shared Resource. Each experiment was repeated at least three times.
2.10 LysoTracker studies for detecting LMP
2.10.1 Confocal microscopy
12 × 104 cells were plated in Ibidi µ-Slide eight-well glass bottom coverslips and treated with vehicle, 1 μM abemaciclib, 5 μM HCQ, or the combination for 24 h. Live cells were treated with LysoTracker Green DND-26 (Cell Signaling Technology) and Hoechst 33342 (Cell Signaling Technology) according to the manufacturer's protocol and imaged with a Leica SP8 Confocal microscope through the Microscopy and Imaging Shared Resource.
2.11 CD68 staining
Cells were washed twice with PBS and resuspended in Fixation and Permeabilization solution (BD Biosciences #554722) and incubated at 4°C for 20 min. Cells were washed twice with 1x Perm/Wash buffer (BD Biosciences #554723) and resuspended in 1x Perm/Wash buffer. TruStain FcX (BioLegend #101319) was added and cells were incubated on ice for 15 min. CD68 antibody (Biolegend #137013) was added and cells were incubated on ice for 30 min. Cells were washed with 1x Perm/Wash buffer and resuspended in Maxpar Cell Staining buffer (Fluidigm Corp #201068) for analysis by the Flow Cytometry Shared Resource.
2.12 Analysis of TFEB gene mutation with cBioPortal
Presence of TFEB gene mutations in breast cancer was determined by using the cBio Cancer Genomics Portal (http://cbioportal.org).23 A mutational analysis was performed in 7298 breast cancer samples from nine studies: The Metastatic Breast Cancer Project (Provisional, December 2021), Proteogenomic landscape of breast cancer (CPTAC, Cell 2020), The Metastatic Breast Cancer Project (Archived, 2020), Metastatic Breast Cancer (INSERM, PLoS Med 2016), Breast Invasive Carcinoma (TCGA, Firehose Legacy), Breast Invasive Carcinoma (TCGA, Cell 2015), Breast Invasive Carcinoma (TCGA, PanCancer Atlas), Breast Cancer (METABRIC, Nature 2012 & Nat Commun 2016) and Breast Invasive Carcinoma (TCGA, Nature 2012).
2.13 Statistical analysis, determination of synergistic interactions and overall survival (OS) analysis
Graphpad Prism 9 was used for statistical analysis. Proliferation, tumor growth rate, cell cycle, LysoTracker imaging, apoptosis, necrosis, and Cyto-ID assays were analyzed using 2-way analysis of variance (ANOVA) tests. The SynergyFinder R package was used to determine Bliss scores. A score > 10 indicates a synergistic interaction, 10 to −10 indicates additivity and <−10 indicates antagonistic interaction.24 Unpaired t-tests were used to analyze the basal and TFEB siRNA invasion and migration assays, tumor necrosis, CD68 expression. One-way ANOVA was used to analyze migration and invasion assays with drug treatment. p values of <0.05 were considered significant. OS data is presented as Kaplan−Meier plots and were generated through Kaplan−Meier plotter (https://kmplot.com/analysis/)25 by selecting breast cancer RNA-seq data sets and querying for TFEB gene and applying the filters for OS, ER+ and lymph node positive that resulted in 914 samples.
3 RESULTS
3.1 Metastatic potential of breast cancer cells is increased with abemaciclib resistance
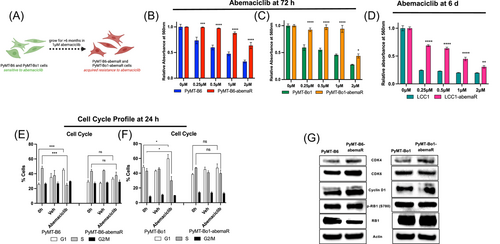
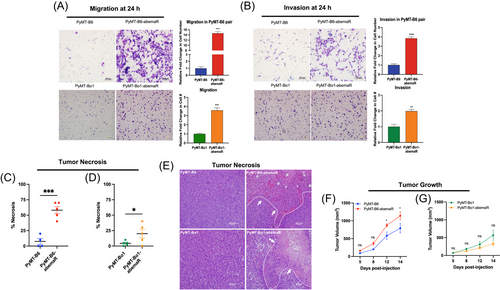
Murine MMTV-PyMT parental cells, PyMT-B6 and its derivative, PyMT-Bo1,18 were used to generate PyMT-B6-abemaR and PyMT-Bo1-abemaR, respectively, by culturing cells in 1μM abemaciclib over 6 months (Figure 1A). The basal growth rate of resistant cells remained comparable to parental sensitive cells (Supporting Information: Figure S1A,B). To determine cellular sensitivity to abemaciclib, we measured cell proliferation with vehicle or increasing concentrations of abemaciclib over 72 h (Figure 1B,C). PyMT-B6-abemaR and PyMT-Bo1-abemaR cells showed a significant (p < 0.05) decrease in sensitivity to abemaciclib at 0.25-2μM compared to vehicle treated cells. IC50 values for abemaciclib for all cell lines used in this study are included in Supporting Information: Table S1. The aforementioned method was also used to generate abemaciclib resistant ER+ LCC1 cells, which are MCF7 derivates that are not sensitive to estrogen.19 Similarly, LCC1-abemaR cells showed a significant decrease in sensitivity to abemaciclib at 0.25−2 μM (Figure 1D). Moreover, PyMT-B6-abemaR and PyMT-Bo1-abemaR showed significant cross-resistance to both palbociclib and ribociclib (Supporting Information: Figure 1SC-F). PyMT-B6-abemaR and PyMT-Bo1-abemaR cells failed to arrest in the G1 phase of the cell cycle while sensitive PyMT-B6 and PyMT-Bo1 cells showed an increase in G1 cell cycle arrest as expected (Figure 1E,F). For PyMT-B6 and PyMT-Bo1 cells, both abemaciclib resistant and parental cells are ER+ and express low levels of progesterone receptor (PR) and human Epidermal Growth Factor Receptor 2 (HER2/ERBB2) (Supporting Information: Figure S1G). Abemaciclib targets CDK4/6 that complexes with cyclin D1 to phosphorylate and inactivate RB1.26 Protein levels of CDK4, CDK6 and cyclin D1 were notably increased in resistant cells while phospho-RB1 (S780) levels remained unchanged compared to sensitive cells (Figure 1G). Since both PyMT-B6 and PyMT-Bo1 cells are highly metastatic,18 we tested whether resistance to abemaciclib altered their metastatic potential using in vitro migration and invasion assays. Interestingly, both PyMT-B6-abemaR and PyMT-Bo1-abemaR cell lines were significantly more migratory and invasive than their respective parental sensitive cells (Figure 2A,B). A significant increase in migration and invasion was also observed in LCC1-abemaR cells relative to parental LCC1 cells (Supporting Information: Figure S1H and S1I). Moreover, pathological evaluation of primary tumors following orthotopic injection of sensitive or resistant PyMT-B6 and PyMT-Bo1 cells showed significant increase in necrotic tissue and immune cell infiltration (Figure 2C−E) while the basal growth rates of the sensitive and resistant tumors were comparable (Figure 2F,G). Murine PyMT-B6 and PyMT-Bo1 are metastatic cell lines and are used as models of luminal B breast cancer models,18 while human LCC1 cells are used as a model of aromatase inhibitor resistance.19 In all three cell lines, abemaciclib resistance was associated with increased migratory and invasive potential. Together, these data show that breast cancer cells that acquire resistance to abemaciclib also gain increased metastatic potential.
3.2 Tumorigenic lysosomal proteins are increased in abemaciclib resistant cells
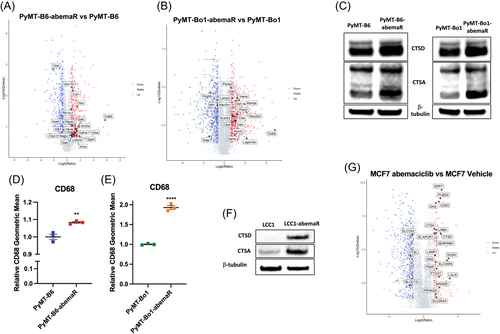
To evaluate proteomic changes that confer resistance to abemaciclib, we conducted mass spectrometry based global proteomics analyses with sensitive PyMT-B6 and PyMT-Bo1 and their respective PyMT-B6-abemaR and PyMT-Bo1-abemaR cells. In total, >5400 proteins were identified and quantified across samples. Among them, hundreds of proteins were significantly changed between sensitive and resistant cells. Of note, very striking changes were observed for many lysosomal proteins (Figure 3A,B; Supporting Information: Table S2 and S3), especially those with tumorigenic qualities such as CTSA, CTSD, cathepsin Z, and CD68 (lysosomal/endosomal-associated membrane glycoprotein LAMP4).27, 28 Western blot analysis for CTSD and CTSA (Figure 3C) and flow cytometry for CD68 (Figure 3D,E) confirmed increased levels of these proteins as detected in the proteomics analysis in resistant cells compared with sensitive cells. An increase in lysosomal proteins CTSD and CTSA is also observed in LCC1-abemaR cells compared to sensitive parental cells (Figure 3F). Previously, it was shown that abemaciclib treatment induced an increase in lysosome-based vacuole formation in treatment-naïve lung cancer cells.9 To determine if abemaciclib induced lysosomal proteins in sensitive breast cancer cells, we treated human MCF7 cells with 1 μM abemaciclib for 72 h and performed quantitative proteomics (Figure 3G). Indeed, up to 3% of all significantly changed proteins in abemaciclib treated MCF7 cells were lysosomal proteins, including CSTD and CTSA (Supporting Information: Table S4). Proteins upregulated with abemaciclib treatment that are shared with those increased in abemaciclib resistant cells are bolded in Supporting Information: Table S4. Common proteins upregulated in abemaciclib resistant PyMT-B6 and PyMT-Bo1 cells are bolded in each table. Together, these data show that lysosomal proteins are remarkably increased with abemaciclib treatment and abemaciclib resistant cells retain tumorigenic lysosomal proteins.
3.3 Lysosomal destabilizers resensitize resistant cells to CDK4/6 inhibitors and reduce metastatic potential
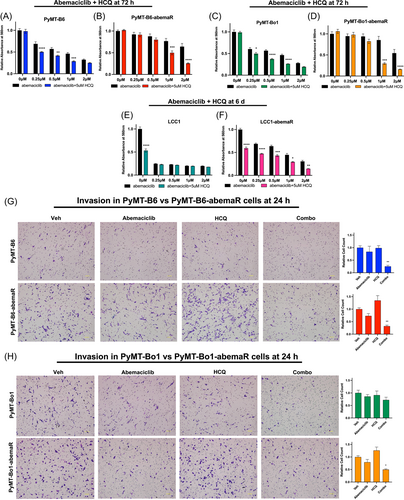
To evaluate the role of lysosomes in antiproliferative effects of abemaciclib, we treated sensitive and resistant cells with known destabilizers of lysosomal functions such as Baf A1, ammonium chloride (NH4Cl) and HCQ. Baf is a V-ATPase inhibitor that prevents lysosomal acidification and autophagosome-lysosome fusion,29 NH4Cl and HCQ are lysosomotropic agents that become protonated and trapped within lysosomes.30 HCQ is a clinical grade anti-malarial drug that is also used as an anticancer drug for its ability to disrupt lysosome-autophagosome fusion.31 Since Baf is highly toxic to all cells within 24 h, a dose response with increasing concentrations of Baf was done within 15 h (Supporting Information: Figure S2A,B) while dose responses with NH4Cl and HCQ were done for 72 h. Abemaciclib resistant cells were more sensitive to Baf compared to sensitive cells in both PyMT-B6 and PyMT-Bo1 pairs. On the other hand, dose-responses with NH4Cl and HCQ showed increased sensitivity in abemaciclib sensitive cells compared to resistant cells (Supporting Information: Figure S2C-F). Next, we combined a single dose of Baf, NH4Cl or HCQ, based on efficacy (~IC50) as single agent in sensitive cells, with increasing doses of abemaciclib (Supporting Information: Figure S3A-H, Figure 4A−D). To determine synergistic interaction between abemaciclib and HCQ, we calculated Bliss synergy scores24 and showed that these drugs interact synergistically, with scores > 10, in both sensitive and resistant PyMT-B6 and PyMT-Bo1 cells (Supporting Information: Figure S3I-J). Notably, abemaciclib and HCQ were more synergistic in resistant cells at doses of abemaciclib > 1 μM in both cell lines. Resistant LCC1-abemaR cells are also re-sensitized to abemaciclib with the addition of HCQ (Figure 4E,F). PyMT-Bo1 cells were also made resistant to palbociclib in the same manner as the abemaciclib resistant cells (Supporting Information: Figure S4A). Both palbociclib sensitive and resistant PyMT-Bo1 cells were further sensitized to palbociclib with the addition of 5 μM HCQ (Supporting Information: Figure S4B,C). Also, sensitive PyMT-Bo1 cells and cells made resistant to ribociclib (Supporting Information: Figure S4D) experienced reduced proliferation when ribociclib was combined with HCQ (Supporting Information: Figure S4E,F). Since PyMT-B6-abemaR and PyMT-Bo1-abemaR cells showed increased metastatic potential compared with parenal cells (Fig. 2A,B), we asked if combination of abemaciclib and HCQ can change their migratory or invasive potential. Results from in vitro migration assays (Supporting Information: Figure S5A,B) showed that while abemaciclib alone inhibited cell migration in PyMT-B6, PyMT-B6-abemaR and PyMT-Bo1-abemaR cells, the combination of HCQ and abemaciclib is significantly more effective in inhibiting migration in these cells. Though, abemaciclib alone or the combination of HCQ and abemaciclib did not change migration in PyMT-Bo1 cells. Results from invasion assays (Figure 4G,H) showed that combination of abemaciclib and HCQ significantly inhibited invasion in both sensitive PyMT-B6 and resistant PyMT-B6-abemaR cells as well as PyMT-Bo1-abemaR cells compared to vehicle alone. Notably, in PyMT-Bo1 cells, neither treatment with monotherapies nor the combination affected invasive potential of the cells. Thus, inhibition of lysosomal function inhibited migration and invasion potential in both abemaciclib sensitive and resistant variants of PyMT-B6 cells. However, in PyMT-Bo1 cells, migration and invasion was significantly inhibited only in the resistant PyMT-Bo1-abemaR cells. Together, these data highlight an essential role of lysosomes in promoting migratory and invasive potential in metastatic breast cancer cells. Furthermore, these data also show that targeting lysosomes can inhibit migratory and invasive potential in abemaciclib resistant cells.
3.4 Abemaciclib resistant cells are more susceptible to lysosomal membrane permeabilization than sensitive cells
Lysosomes are acidic single membrane organelles that contain > 50 different acid hydrolases. Induction of LMP leads to lysosome dependent cell death.32, 33 To asses LMP in abemaciclib sensitive and resistant cells, we incubated PyMT-B6, PyMT-B6-abemaR, PyMT-Bo1 and PyMT-Bo1-abemaR cells with LysoTracker Green and evaluated lysosomal staining with live confocal microscopy. Treatment with 1 μM abemaciclib induced robust accumulation of LysoTracker within lysosomes compared to vehicle control. Treatment with 5 μM HCQ did not show a marked difference in LysoTracker staining compared to control. However, treatment with combination of abemaciclib and HCQ diminished LysoTracker accumulation (Figure 5A,B). Therefore, addition of HCQ along with abemaciclib disabled the formation of intact lysosomes as observed with abemaciclib treatment alone. Since LMP can induce cell death through a variety of pathways 33, we compared the percentage of cells undergoing apoptosis using annexin V assays (Figure 6A,B) in PyMT-B6 and PyMT-Bo1 abemaciclib sensitive and resistant pairs under the different treatment conditions. We found that treatment with abemaciclib alone or the combination of abemaciclib and HCQ induced a significant but modest increase (~10%) in apoptosis in sensitive PyMT-B6 cells compared with vehicle treatment. In PyMT-B6-abemaR cells, a significant but modest (~5%) increase in apoptotic cells was present only with the combination treatment compared with vehicle. In PyMT-Bo1 cells, abemaciclib alone or the combination of abemaciclib and HCQ induced a significant and robust increase (40%) in apoptotic cells, while abemaciclib alone or in combination with HCQ induced a relatively lesser effect (20%) compared to vehicle in PyMT-Bo1-abemaR cells. Treatment with HCQ alone did not show an increase in apoptotic cells in any of the four cell lines. Next, we determined autophagy induction by measuring accumulation of autophagosomes in both sensitive and resistant cells (Figure 6C,D). Following treatment with either abemaciclib alone or the combination of abemaciclib and HCQ, there was approximately a four- and two-fold significant induction of autophagosomes in PyMT-B6 cells and PyMT-B6-abemaR cells, respectively. In PyMT-Bo1 cells, induction of autophagosomes was 2-fold with abemaciclib alone or in combination with HCQ. No significant differences were seen with any treatment conditions in PyMT-Bo1-abemaR cells. Serum deprived starvation was used as positive control in detecting autophagosomes. Interestingly, we noticed a varied response between sensitive and resistant cells to starvation, although in all cells, starvation induced increased accumulation of autophagosomes. To further confirm the induction of cell death pathways, we conducted Western blot analysis with known markers apoptosis and autophagy (Figure 6E). In both PyMT-B6 and PyMT-Bo1 sensitive cells, abemaciclib alone, or more markedly, abemaciclib and HCQ treatment induced an increase in cleaved PARP1 (poly(ADP-ribosyl)transferase), a known marker of apoptosis.34 In contrast, cleaved PARP1 was not increased in PyMT-B6-abemaR and PyMT-Bo1-abemaR cells with abemaciclib alone or in combination with HCQ compared to vehicle. To measure autophagic flux, we examined protein levels of p62 (SQSTM1), an autophagosome cargo marker that decreases in active autolysosomes, and LC3B-II, a marker that increases with formation and enlargement of autophagosomes.35 Abemaciclib alone decreased p62 levels and increased LC3B-II compared to vehicle in sensitive PyMT-B6 cells only. HCQ alone induced a modest increase in LC3B-II without a concomitant decrease in p62 compared to vehicle in all cell lines suggesting incomplete autophagy. Combination of HCQ and abemaciclib increased LC3B-II levels with simultaneous decrease in p62 in sensitive PyMT-B6 and PyMT-Bo1 cells but not in respective resistant cells. Considering the relatively subdued induction of apoptosis or autophagy in resistant cells, we examined the protein levels of BCL2 (B cell lymphoma 2), a well-characterized inhibitor of multiple death pathways, including apoptosis, necrosis, and autophagy.36, 37 Interestingly, BCL2 levels were increased in resistant cells compared to sensitive cells in both PyMT-B6 and PyMT-Bo1 cell lines (Figure 6F) and remained detectable following treatment with HCQ, abemaciclib or the combination in resistant cells. Furthermore, BCL2 protein was markedly increased in LCC1-abemaR cells compared to LCC1 cells (Figure 6G). Since increased levels of BCL2 protein were detected in abemaciclib resistant cells, we tested the efficacy of BCL2 inhibitors, venetoclax (VNX) (Figure 6H) and obatoclax (OBT) (Supporting Information: File 9, Figure S6A) in PyMT-Bo1 and PyMT-Bo1-abemaR cells. Based on these dose–response studies, we used the IC50 concentrations for VNX (7 μM) and OBT (15 nM), in PyMT-Bo1 cells, to test the efficacy of triple combination of HCQ, abemaciclib and a BCL2 inhibitor in abemaciclib sensitive and resistant cells. Combination of HCQ, abemaciclib and VNX had the same inhibitory effect as HCQ and abemaciclib in PyMT-Bo1 cells (Figure 6I). However, the combination of HCQ, abemaciclib and VNX significantly decreased cell proliferation compared to the combination of HCQ and abemaciclib in PyMT-Bo1-abemaR cells (Figure 6J). Combination of HCQ, abemaciclib and OBT significantly decreased cell proliferation compared to the combination of HCQ and abemaciclib in both PyMT-Bo1 and PyMT-Bo1-abemaR cells (Supporting Information: Figure S6B,C). VNX specifically inhibits BCL2 while OBT is a pan-BCL2 inhibitor and can inhibit multiple members of the BCL2 family, including BCL2L1 (BCLxL) and MCL1.38 Together, these data show that increased levels of BCL2 contributes to acquired resistance to abemaciclib, and supports the use of anti-BCL2 drugs as a therapeutic option in acquired resistance to CDK4/6i.
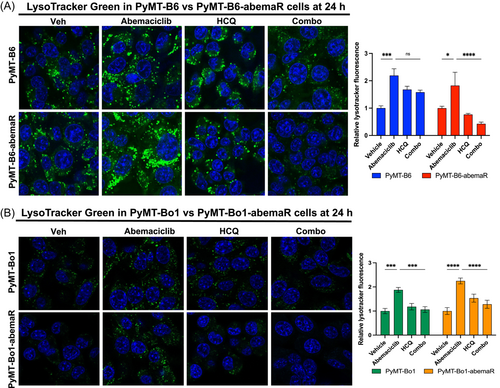
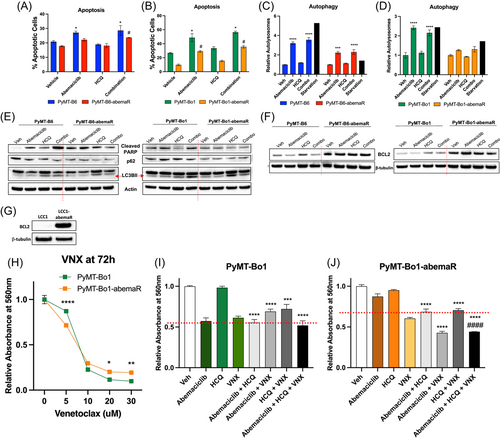
To determine whether combination of abemaciclib and HCQ affected cell cycle, we analyzed cell cycle profiles in sensitive and resistant pairs of PyMT-B6 and PyMT-Bo1 cells. As expected, treatment with abemaciclib alone induced G1 cell cycle arrest in both sensitive cells, PyMT-B6 and PyMT-Bo1, however, combination of abemaciclib and HCQ did not further augment G1 cell cycle arrest in sensitive cells (Figure 7A−D). In resistant PyMT-B6-abemaR and PyMT-Bo1-abemaR cells, neither abemaciclib alone or the combination of abemaciclib and HCQ affected cell cycle profiles compared to vehicle control. Collectively, these data show that combination of abemaciclib and HCQ induce LMP in abemaciclib resistant cells, however, apoptosis, autophagy or cell cycle arrest are not robustly induced compared to sensitive cells.
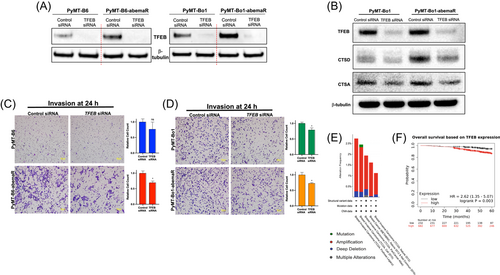
3.5 Genetic alteration of TFEB is associated with tumorigenesis in ER+ breast cancer
TFEB is a master regulator of a number of lysosomal and autophagy proteins. Since lysosomal proteins were deregulated in abemaciclib resistant cells, we analyzed TFEB protein levels in abemaciclib sensitive and resistant cells. Western blot analysis showed increased TFEB protein levels in PyMT-B6-abemaR and PyMT-Bo1-abemaR cells compared to their sensitive parental cells (Figure 7A). siRNA-mediated knockdown of TFEB reduced proliferation in both sensitive and resistant PyMT-B6 and PyMT-Bo1 cells after 72 h (Supporting Information: Figure S8A-D). In PyMT-Bo1-abemaR cells, knockdown of TFEB showed decrease in CTSD compared with PyMT-Bo1 sensitive cells (Figure 7B). Knockdown of TFEB with siRNA also showed a significant decrease in invasive potential of PyMT-B6-abemaR and PyMT-Bo1-abemaR cells (Figure 7C,D). TFEB has previously been reported to affect invasion but not proliferation in human squamous carcinoma cells.39 Next, we analyzed 7,298 invasive breast cancer patient sample data curated by cBioportal, and found amplifications, deletions and other mutations in 2% of all cases (Figure 7E). Furthermore, analysis of OS in ER+ breast cancer patients showed that increased TFEB expression correlated with lower OS (HR = 2.62 [1.35−5.07]; p = 0.003) (Figure 7F). Together, these data show that increased or genetically altered TFEB expression in breast cancer correlated with increased tumorigenesis in a subset of breast cancer.
4 DISCUSSION
CDK4/6i are used in combination with antiestrogens as the first-line treatment of ER+ metastatic breast cancer, and abemaciclib in particular is used in the adjuvant setting for patients with early, high-risk disease.4 Although CDK4/6i successfully extend progression-free survival, resistance is a common problem, and currently, there are no FDA approved drugs to specifically treat CDK4/6i resistant breast cancer.40 To this point, research into resistance had focused on implicating growth factors or cell cycle proteins,41 however, efficacy of these agents remain clinically unconfirmed. Recently, lysosomes have been implicated in a non-apoptotic mode of cell death in response to abemaciclib and as a mode of inherent resistance to palbociclib in TNBC.9, 10 Similarly, we show that in sensitive MCF7 cells, abemaciclib induced an increase of lysosomal proteins (Figure 3G; Supporting Information: Table S3) compared to vehicle treatment. Hence, abemaciclib triggers activation of cellular mechanisms that increase lysosomal proteins in ER+ abemaciclib sensitive breast cancer cells. Furthermore, we show that breast cancer cells that acquire resistance to abemaciclib are more metastatic (Figure 2A,B; Supporting Information: Figure 1S H,I) and contain higher levels of protumorigenic lysosomal proteins (Figure 3A,B). Therefore, targeting the lysosomal pathway is a plausible therapeutic strategy in advanced ER+ metastatic breast cancer.
Lysosomes are primarily known for catabolic turnover of various macromolecules but these versatile organelles are also regulators of cell proliferation and promoters of metastasis.17, 42 Lysosomes are controlled by the microphthalmia/transcription factor E (MiT/TFE) family of transcription factors consisting of MITF, TFE3, TFEC, and TFEB.43 TFEB has many functions, but regulation of lysosomal-autophagy genes is one that can directly contribute to metastasis.44 In abemaciclib resistant breast cancer models, while we do not see changes in lysosomal number with vehicle alone or abemaciclib (Figure 5A,B), levels of a number of lysosomal proteins were significantly changed (Figure 3), including increase in CTSD. Furthermore, siRNA mediated knockdown of TFEB decreased CTSD and CTSA proteins in resistant cells (Figure 7B). Thus, increased levels of TFEB in resistant cells could augment levels of protumorigenic lysosomal proteins such as CTSD and CTSA, which can drive tumorigenesis. Through analyses of publicly available human breast cancer data, we show that TFEB mutations, particularly amplification, are present in a subset of invasive breast cancer cases, and increased TFEB gene expression correlated with decreased OS in ER+ breast cancer cases (Figure 7D,E). Evaluation of TFEB protein levels in 100 breast cancer patients with small tumors ( < 2 cm) showed high TFEB expression in one-fourth of the cases.45 Increased TFEB was found to be a prognostic variable and also positively correlated with increase in the autophagosome geneLC3A levels in this cohort. Therefore, it is possible that in a subset of breast tumors that harbor increased TFEB levels, treatment with CDK4/6i will lead to poor clinical outcome, and targeting TFEB-mediated pro-survival pathways could improve efficacy of CDK4/6i in these cases.
HCQ is widely used as an inhibitor of the autophagy-lysosomal pathway.46 HCQ is a weak base that accumulates in acidic compartments of cells and tissue. HCQ can disrupt lysosomal function, autophagy, membrane integrity as well as transcription and signaling.47 Like HCQ, abemaciclib is lysosomotropic10 and the combination of these two drugs disrupts lysosomal integrity by inducing LMP, and downstream death, in resistant cells (Figure 5A,B). The discrepancy in apoptosis levels measured by annexin-V assay versus Western blot analysis could be due to PARP-independent induction of apoptosis following treatment with abemaciclib and HCQ in resistant cells (Figure 6A,B,E). Experiments to determine this are currently underway. However, it is noteworthy that cathepsin mediated regulation of PARP1 can promote tissue survival by shifting the balance of cell death programs, so it is possible that the upregulation of cathepsins in resistant cells could be responsible for the altered apoptotic signaling under basal conditions.48 We noted a reduction in death in resistant cells compared to sensitive cells, and found an upregulation of BCL2 in these cells (Figure 6). Recently, two trials have begun to test the efficacy of HCQ or BCL2 inhibitors with CDK4/6i in ER+ breast cancer. The ABBY trial is investigating the efficacy of adding HCQ, as an autophagy inhibitor, to abemaciclib in CDK4/6 inhibitor-naïve patients to prevent survival of senescent breast cancer cells disseminated in the bone.49 The PALVEN trial is designed to test the safety of palbociclib, letrozole, and a BCL2 inhibitor, VNX, as adding a BCL2 inhibitor to CDK4/6i triggers higher levels of apoptosis in cancer cells.50 These studies provide evidence that adding targeted therapies, which disrupt survival signaling that inhibit cell death pathways, to CDK4/6i can increase their efficacy. Future studies will investigate the effects of combining abemaciclib and other lysosome destabilizing drugs on tumor growth and metastasis using in vivo models of ER+ breast cancer.
5 CONCLUSIONS
CDK4/6i along with antiestrogen therapy has become the gold standard of treatment choice for ER+ metastatic breast cancer. Since CDK4/6i are used in treatment of advanced breast cancer, one of the greatest clinical challenges has been to unravel mechanisms of CDK4/6i resistance from that of antiestrogen resistance.51 To date, RB1 loss or mutation, modification of CDK4,52 CDK6 gene/protein levels,53 or genomic alteration of different of signaling pathways54 have been proposed in CDK4/6i resistance in small patient subsets. However, these molecular modifications cannot explain CDK4/6i resistance observed in larger groups of breast cancer cases, and therefore, biomarkers are urgently needed to identify therapeutic strategies to treat advanced disease. General treatment duration with CDK4/6i typically last over many months. Eventually, all treated tumors will develop resistance.1 At the cellular level, treatment with CDK4/6i induced lysosomes within minutes9, 10; (Figure 5), however, the long-term effect remains unknown. In this study, we showed that CDK4/6i resistant ER+ breast cancer cells are more metastatic and comprise increased levels of protumorigenic lysosomal proteins, and lysosomal targeting drugs can be combined with CDK4/6i to inhibit growth of resistant cells. Further studies are warranted to establish whether interfering with lysosomal function can inhibit growth of primary tumors and metastasis in vivo.
AUTHOR CONTRIBUTIONS
Erin R. Scheidemann, Diane M. Demas, Chunyan Hou, Junfeng Ma, Wei He, Gaurav Sharma, Eric Schultz, Katherine N. Weilbaecher and Ayesha N. Shajahan-Haq participated in the experimental design, statistical analysis, and interpretation of results. Erin R. Scheidemann, Diane M. Demas, Chunyan Hou, Junfeng Ma and Ayesha N. Shajahan-Haq carried out the experiments. Erin R. Scheidemann and Ayesha N. Shajahan-Haq drafted and revised the manuscript. Erin R. Scheidemann, Junfeng Ma, Katherine N. Weilbaecher and Ayesha N. Shajahan-Haq critically revised the manuscript for important intellectual content. All authors read and approved the final manuscript.
ACKNOWLEDGMENTS
We thank Idalia Cruz for technical assistance with the in vivo studies, and Dr. Susana Galli (veterinary pathologist) for pathological evaluation of the murine primary tumor samples. We also thank the Georgetown Breast Cancer Advocates (GBCA) for a patient's perspective for the translational aspect of our study. Funding support for this study is in part from Georgetown University Medical Center and a METAvivor Translational Research Award to ANS-H. Technical services were provided by the following shared resources at Georgetown University Medical Center: Tissue Culture, Flow Cytometry, Animal Models and Mass Spectrometry and Analytical Pharmacology Shared Resource that were funded through Public Health Service award P30-CA-51008 (Lombardi Comprehensive Cancer Center Support Grant). The instrument Orbitrap Lumos Tribrid mass spectrometer was partially supported by Dekelbaum Foundation.
CONFLICT OF INTEREST STATEMENT
G. S. and E. S. hold equity in Ocean Genomics, Inc. The remaining authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.




