Synthesis of Passerini-3CR Polymers and Assembly into Cytocompatible Polymersomes
Abstract
The versatility of the Passerini three component reaction (Passerini-3CR) is herein exploited for the synthesis of an amphiphilic diblock copolymer, which self-assembles into polymersomes. Carboxy-functionalized poly(ethylene glycol) methyl ether is reacted with AB-type bifunctional monomers and tert-butyl isocyanide in a single process via Passerini-3CR. The resultant diblock copolymer (P1) is obtained in good yield and molar mass dispersity and is well tolerated in model cell lines. The Passerini-3CR versatility and reproducibility are shown by the synthesis of P2, P3, and P4 copolymers. The ability of the Passerini P1 polymersomes to incorporate hydrophilic molecules is verified by loading doxorubicin hydrochloride in P1DOX polymersomes. The flexibility of the synthesis is further demonstrated by simple post-functionalization with a dye, Cyanine-5 (Cy5). The obtained P1-Cy5 polymersomes rapidly internalize in 2D cell monolayers and penetrate deep into 3D spheroids of MDA-MB-231 triple-negative breast cancer cells. P1-Cy5 polymersomes injected systemically in healthy mice are well tolerated and no visible adverse effects are seen under the conditions tested. These data demonstrate that new, biodegradable, biocompatible polymersomes having properties suitable for future use in drug delivery can be easily synthesized by the Passerini-3CR.
The increasing need for complex functional polymers in challenging biomedical applications requires chemistries which enable preparation of materials with highly defined macromolecular architectures, but without complex, costly, and unsustainable synthetic procedures. Multicomponents reactions offer potential solutions to these challenges, due to their high atom economy, degree of versatility, and ease of synthetic protocols.[1] The Passerini three component reaction (Passerini-3CR) is one of the most well-established of these reactions, combining in a one-pot procedure a carboxylic acid, an isocyanide, and an oxo-component (aldehyde or ketone) to synthesize an α-acyloxycarboxamide.[2] The versatility of the Passerini-3CR has, for instance, been exploited for the synthesis of monomers,[3] multifunctional RAFT agents,[4] star-shaped unimolecular micelles,[5] and linear polymers.[6] The combination of different monomers through a step-growth polymerization mechanism allows the synthesis of a variety of tunable, biodegradable, and biocompatible polyesters, and polyamides with functional side chains.[7] Accordingly, there are many potential biomedical applications for Passerini-type polymers, with an area of particular interest being the use of Passerini chemistries to generate drug delivery vehicles. This is because the degradability of the polyester linkages, combined with the potential for tuning functionality and architecture for pro-drug formation or drug encapsulation, overcome many of the problems inherent to other polymers investigated as therapeutic carriers. However, there have been relatively few studies to date of Passerini-type polymers in which some key parameters essential to their practical application for therapeutic delivery have been evaluated. These include their ability to self-assemble into structures suitable for drug encapsulation, such as micellar-like nanoparticles and/or polymersomes, or their compatibility with target cell types and their organ accumulation following systemic injection. Here, the focus is on Passerini chemistries to generate polymersomes, as these are versatile drug carriers, which can encapsulate both hydrophobic molecules within the bilayer and hydrophilic molecules in the aqueous core, while also reducing off-target toxicity.[8] Polymersomes derived from other chemistries have been widely explored for drug delivery applications,[9] but their translation to date has been hampered by the lack of reproducibility of the synthetic and formulation procedures.[10] The authors set out therefore to use the simplicity of the Passerini-3CR to synthesize amphiphilic copolymers that could assemble in an easy and reproducible way into polymersomes. The authors report here the synthesis, cell internalization, and cytocompatibility of Passerini polymersomes, and their transport in 3D cell cultures and organ accumulation in a mouse model.
The Passerini-3CR protocol was adapted from a previous report,[11] where this multicomponent reaction was performed as a step-growth polymerization and with good control over the polymer molecular weight and architecture. The synthesis of the linear amphiphilic diblock copolymer P1 through the Passerini-3CR was based on the introduction of mPEG-COOH (4) as irreversible chain transfer agent (ICTA) in a polymerization reaction involving AB-type monomers (3) (combination of carboxylic acid and aldehyde; 1:60 molar ratio) and excess of tert-butyl isocyanide (Figure 1A). A total of 41 AB monomer units were attached per ICTA-PEG chain in P1 copolymer, corresponding to 68% of monomer conversion. The collected P1 copolymer molecular weight was calculated by 1H NMR, using the signal correlation between the mPEG CH3 singlet and the polymer repeating units peaks of the amide (NH) and the adjacent and chiral CHi (see Supporting Information for details). The calculated molecular weight was found to be 19 700 Da. The SEC analysis showed a molar mass Mn of 19 700 Da and dispersity (Đ) of 1.44, which is in agreement with previous reports for the polymers synthesized by Passerini-3CR.[12] The reproducibility and the versatility of the Passerini-3CR syntheses were assessed by reacting and varying the molar ratio of the AB-type monomer with different ICTAs and tert-butyl isocyanide, leading to the preparation of P2, P3 and P4 Passerini copolymers. mPEG-COOH (5 kDa) (4) was reacted with the AB-type monomer with molar ratio of 1:60 (P1) and 1:50 (P2), respectively, leading to an average percentage of AB monomer attached per ICTA chain of 69 ± 1.41% (see Supporting Information). The commercially available methoxy polyethylene glycol 5000 propionic acid was reacted with the AB-type monomer in a molar ratio of 1:30 leading to a percentage of AB monomer attached per PEG chain of 70%, which is in agreement with what was observed for P1 and P2 Passerini diblock copolymers. Therefore, the Passerini-3CR was found to be a reproducible polymerization, which leads to control the length of the hydrophobic block and, thus, the copolymer amphiphilicity. Finally, the reactivity of the AB-type monomer was assessed by performing the Passerini-3CR with 10-undecenoic acid as previously reported by Oelmann et al.[11] In this case, all the AB-type monomers were consumed during the Passerini-3CR leading to the synthesis of the hydrophobic P4 polymer.
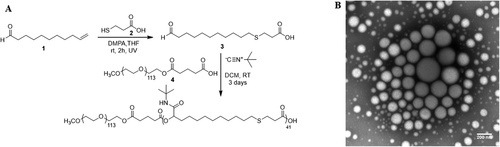
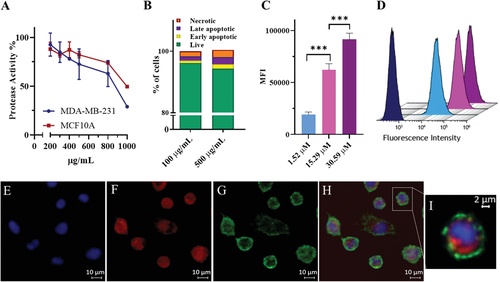
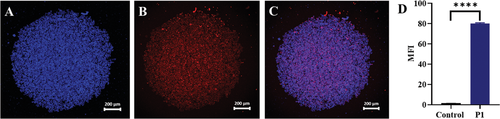
The hydrophobicity of P1 Passerini diblock copolymer was tuned to be 74%, with a hydrophilic weight fraction of 0.26. Therefore, as reported by Eisenberg,[13] Mallik,[14] and Zentel et al.,[15] the amphiphilicity suggested the copolymer could self-assemble into vesicles with double-layers within an aqueous environment.[16] P1 polymersomes were obtained in phosphate buffer (150 mm) with an average diameter intensity size of 145 ± 45 nm and a zeta potential of −5 ± 7 mV. The morphology of the P1 superstructures and the formation of polymersomes was assessed by transmission electron microscopy (TEM), as shown in Figure 1B and Supporting Information. The P1 polymersomes morphology was further assessed by loading a water-soluble molecule in the core of the vesicles. Therefore, doxorubicin-loaded P1 polymersomes (P1DOX; 127 ± 33 nm) were formulated in water containing doxorubicin hydrochloride at pH 6. The non-encapsulated drug was removed using a Sephadex G-25 column and the drug loading (0.94 ± 0.003%) and encapsulation efficiency (2.35 ± 0.003%) were quantified by HPLC analysis (see Supporting Information). Since it is intended ultimately to use these polymers in oncology drug delivery applications, the effects of P1 copolymers on non-cancerous MCF10A human mammary epithelial cells and on MDA-MB-231 triple negative breast cancer cells were investigated. The CellTiter-FluorCell Viability Assay was performed in order to measure the activity of protease enzyme in cells populations treated with P1 copolymer in a concentration range of 200–1000 µg mL−1 for 48 h. The assay protocol was first optimized, as reported in Supporting Information. The fluorescence was recorded as a measure of the constitutive protease activity within live cells and was considered as a biomarker of cell viability.[5] P1 was found to be well tolerated by MDA-MB-231 and MCF-10A cells at concentrations below 800 µg mL−1. Therefore, the toxicity of P1 was further investigated by performing Annexin-V/PI assays, which were used to discriminate between viable, apoptotic and dead cells using flow cytometry.[17] MDA-MB-231 cells were incubated with P1 polymersomes at 100 µg mL−1 and 500 µg mL−1 in media for 48 h. At the lower concentration (100 µg mL−1) P1 was less toxic as measured by the percentage of live cells (96.18% ± 0.29) compared to the number of live cells when P1 at 500 µg mL−1 was used (94.34% ± 0.65). These data indicated that, for this assay at least, P1 caused only a small reduction in cell viability as deduced from protease activity. The above experiments established the ability of polymer P1 to form polymersomes, which did not adversely affect viability of non-cancerous breast and triple negative breast cancer cells (Figure 2A,B). The versatility of the Passerini copolymer synthesis for installing accessible end-group functionality was exploited for the conjugation of the P1 copolymer carboxylic acid end group with amino Cy5 via amidation, leading to 50% yield of functionalization. The obtained fluorescently labelled copolymer (P1-Cy5) was formulated into polymersomes, again via addition to phosphate buffered aqueous solutions. The P1-Cy5 polymersomes size was measured by DLS and the diameter average intensity size was found to be 125 ± 43 nm (zeta potential −2 ± 6 mV). The authors then evaluated the internalization of the P1-Cy5 polymersomes in 2D monolayer and 3D spheroids of MDA-MB-231 cells by flow cytometry and confocal microscopy. P1-Cy5 polymersomes were incubated in 2D MDA-MB-231 cells monolayers for 4 h at a range of concentrations (1.52, 15.29, and 30.59 µm). Flow cytometry data, as reported by the mean fluorescence intensity (MFI) of cells, was found to increase with the concentration of P1-Cy5 polymersomes (Figure 2C,D). The intracellular location of P1-Cy5 polymersomes was then assessed by confocal microscopy in 2D monolayers and 3D spheroids of MDA-MB-231 cells. Figure 2E–I and Supporting Information show the intracellular accumulation of P1-Cy5 polymersomes in MDA-MB-231 cell 2D monolayers after 4 h incubation. The images suggest that P1 polymersomes were dispersed throughout the cytosol, suggesting potential escape from intracellular compartments following internalization via endocytosis.[18] This is the most common cell entry route for nanomaterials with a size range of 100–200 nm,[19] which are usually engulfed in the cell membrane and, then, are transported by dynamic vesicles from the plasma membrane to the cytoplasm.[18] The ability of P1-Cy5 polymersomes to penetrate through model tumor tissue was assessed using 3D spheroids of MDA-MB-231 cells treated for 16 h with 0.047 µm of P1-Cy5 polymersomes. The confocal images in Figure 3A–C indicate the presence of the Cy5 signal throughout the spheroids, further confirming the ability of the Passerini polymersomes to transport inside the cells both in 2D and 3D. The internalization of the P1-Cy5 polymersomes was quantified in the 3D spheroids, as for the 2D monolayers, by flow cytometry. The MFI data in Figure 3D show how the P1-Cy5 polymersomes were efficiently internalized by the MDA-MB-231 cells in the spheroids.
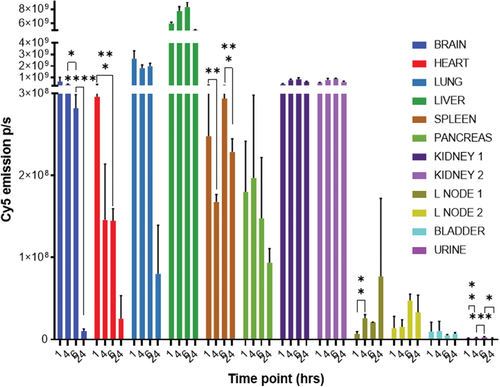
The culminating experiments aimed to assess the pre-requisites for safety in the use of Passerini polymersomes as a drug delivery system. These assays utilized the ‘empty P1-polymersomes, evaluating in the first instance the circulation kinetics and primary organ accumulation of P1-Cy5 polymersomes in healthy mice following injection of the formulation via the tail vein. Within the first 6 h the highest accumulation of P1-Cy5 polymersomes occurred in the lungs and liver (Figure 4). This could be explained by the fact that the lung is, after the heart, the first organ encountered after injection, and the liver is the main sink organ. Thus, the polymersomes might be trapped in the lung capillaries if self-association occurred beyond the diameter of these fine capillaries (>2 µm),[20] or if plasma proteins adsorbed to cause agglomeration. In addition, brain accumulation was seen within 6 h, which might also be explained by absorption of the polymersomes in the blood brain barrier capillaries.[21] However, signals for polymer in both lung and brain decreased very significantly by 24 h, indicating that irreversible aggregation and accumulation due to polymersome instability did not occur. It is thus likely that the accumulation seen at early stages in lungs and brain arose from the larger particles in the polymersome populations, as for these preliminary experiments the authors did not optimize the formulations to be monodisperse. The polymersomes were also found in the kidneys, spleen, and pancreas, again as expected due to their size range of 100–200 nm. The organ showing the highest signal of P1-Cy5 polymersomes was the liver, and this did not diminish significantly over 24 h, suggesting retention and/or prolonged circulation. Furthermore, the liver is the major site of lipoprotein turnover and the 24 h mice image exhibited fluorescence from the polymers in brown adipose tissue (see Supporting Information). It is known that adipose tissue can play a key role in the long-term accumulation and retention of the formulation.[22] The authors did not vary the preparation protocols for the empty P1 polymersomes to optimize their pharmacokinetics, as individual formulations would in any case be required, depending on which drugs are to be encapsulated, and these would inevitably vary in particle size and biodistribution. However, these initial in vivo assays showed that P1-Cy5 polymersomes were well tolerated as the formulation did not produce any apparent adverse effects in the mice after administration.
In this work, an amphiphilic diblock copolymer P1 was synthesized via a Passerini-3CR in a simple one-pot versatile and reproducible procedure. The polymer was also easy to formulate into polymersomes, which were of low toxicity to healthy and cancerous breast cells, and which rapidly entered 2D cell monolayers as well as transported efficiently into 3D spheroids of MDA-MB-231 cells. The polymers were also well tolerated in vivo and were retained in circulatory organs at least up to 24 h in healthy mice, suggesting the potential of these types of Passerini polymersomes to be appropriate as carriers for systemic drug delivery. Future studies will investigate the encapsulation and release of drug cargoes to evaluate the potential of the Passerini polymersomes to act as therapeutic delivery systems.
Experimental Section
Passerini-3CR Polymerization of P1 Amphiphilic Diblock Copolymer
To a vigorously stirred solution of mPEG-COOH (5 kDa) (4) (0.25 g, 0.05 mmol) in 2.4 mL of DCM, AB-type monomer (3) (0.822 g, 3.0 mmol) was added slowly and solubilized. Finally, tert-butyl isocyanide (1.245 g, 15 mmol) was added drop wise without dilution. After stirring for 3 days at room temperature, the polymer was precipitated from ice-cold diethyl ether and the amphiphilic diblock copolymer P1 was obtained as a white solid (0.841 g, yield 89%) and characterized by 1H NMR, 13C NMR, and SEC in THF. The syntheses of AB-type monomer (3) and mPEG-COOH (5 kDa) (4) are reported in Supporting Information.
P1 Copolymer Post-Functionalization with Cy5
P1 (39.87 mg, 1.53 µmol) was solubilized in 0.5 mL of dry DMF. N,N′-dicyclohexylcarbodiimide (0.78 mg, 3.82 µmol) and N-hydroxysuccinimide (0.26 mg, 2.29 µmol) were added to the mixture. The reaction mixture was stirred at room temperature for 12 h and then cyanine 5 amine (1 mg, 1.53 µmol) was dissolved in 0.1 mL of dry DMF and subsequently added. The reaction was left for 72 h. The crude mixture was diluted with 0.6 mL of acetone and the polymer was collected by precipitation in cold diethyl ether. The powder was dried, the polymer was solubilized in 0.5 mL of THF, diluted with 1 mL DI water and then it was directly dialyzed against DI water for 48 h (3.5 kDa cut-off, ThermoScientific). P1-Cy5 polymer was collected after freeze drying (38.1 mg, yield 95.5%). The Cy5 conjugation yield was determined by UV–vis spectroscopy as mentioned in Supporting Information.
P1 and P1-Cy5 Polymersomes Formulation
Passerini diblock copolymers P1 and P1-Cy5 polymersomes were prepared by nanoprecipitation. P1 or P1-Cy5 were dissolved at 5 mg mL−1 concentration in tetrahydrofuran (THF) and added slowly to phosphate buffer pH 7.4 150 mm while stirring, using a syringe pump (flow rate: 0.4 mL min−1) at 1:1 volume ratio. The organic solvent was evaporated overnight in the fume hood and the polymersome sizes (z-average diameter) were measured using a NanoZS instrument (Malvern, UK) at 25 °C (DLS).
Passerini P1 Polymersomes Cytotoxicity: CellTiter-Fluor Cell Viability Assay
The CellTiter-Fluor cell viability assay was performed to determine the cytotoxicity of P1 against MDA-MB-231 and MCF10A cells following the protocol provided by the vendor (Promega, Madison WI).[5] Cells were treated for 48 h with different concentrations of P1 (from 0.2 to 1 mg mL−1). Experiments were made in replicates and repeated on different days. The detailed protocol is reported in Supporting Information.
Passerini P1 Polymersomes Cytotoxicity: Annexin V/PI
The Annexin V/PI assay was performed to determine the cytotoxicity of P1 against MDA-MB-231 cells. The protocol is reported in Supporting Information.
Cellular Uptake Studies
The internalization of P1-Cy5 polymersomes was assessed in 2D monolayers and 3D spheroids of MDA-MB-231 cells for 4 and 16 h incubation, respectively. The cells were imaged by confocal microscopy and the uptake was quantified by flow cytometry. The detailed protocol is reported in Supporting Information.
In Vivo Biodistribution Study
In vivo experiments were performed in order to assess the organ accumulation and the tolerability of P1-Cy5 polymersomes. The experiments were conducted under the UK Home Office License number PPL P435A9CF8. The experiments were conducted following LASA good practice guidelines, FELASA working group on pain and distress guidelines, ARRIVE reporting guidelines and local and national ethical guidelines. Mouse organs were excised and imaged using the IVIS Spectrum imaging system. The detailed protocol is reported in Supporting Information.
Acknowledgements
This work was supported by the Engineering and Physical Sciences Research Council (Grant Number: EP/N006615/1; Grant Name: EPSRC Programme Grant for Next Generation Biomaterials Discovery) and the University of Nottingham. This work was also funded by the Royal Society [Wolfson Research Merit Award WM150086] to CA. The authors thank the Nanoscale and Microscale Research Centre and Prof. Steve Atkinson from the University of Nottingham for providing access to advanced light and scanning microscopy instrumentation and Denise McLean for valuable guidance. The authors also thank Esme Ireson and Paul Cooling for skilled technical assistance, and Sian Rankin-Turner, Elizabeth Hufton, and Carol Turrill for invaluable project support. The authors thank Professor Morgan R. Alexander for helpful discussions and for expert support in the EPSRC Next Generation Biomaterials Discovery Programme Grant.
Data Availability
All relevant data are available from the University of Nottingham's Research Data Management Repository: https://rdmc.nottingham.ac.uk
Conflict of Interest
The authors declare no conflict of interest.




