MuRF1 deficiency prevents age-related fat weight gain, possibly through accumulation of PDK4 in skeletal muscle mitochondria in older mice
Kosuke Sugiura and Katsuya Hirasaka contributed equally to this work.
Abstract
Recent studies show that muscle mass and metabolic function are interlinked. Muscle RING finger 1 (MuRF1) is a critical muscle-specific ubiquitin ligase associated with muscle atrophy. Yet, the molecular target of MuRF1 in atrophy and aging remains unclear. We examined the role of MuRF1 in aging, using MuRF1-deficient (MuRF1–/–) mice in vivo, and MuRF1-overexpressing cell in vitro. MuRF1 deficiency partially prevents age-induced skeletal muscle loss in mice. Interestingly, body weight and fat mass of more than 7-month-old MuRF1–/– mice were lower than in MuRF1+/+ mice. Serum and muscle metabolic parameters and results of indirect calorimetry suggest significantly higher energy expenditure and enhanced lipid metabolism in 3-month-old MuRF1–/– mice than in MuRF1+/+ mice, resulting in suppressed adipose tissue gain during aging. Pyruvate dehydrogenase kinase 4 (PDK4) is crucial for a switch from glucose to lipid metabolism, and the interaction between MuRF1 and PDK4 was examined. PDK4 protein levels were elevated in mitochondria from the skeletal muscle in MuRF1–/– mice. In vitro, MuRF1 interacted with PDK4 but did not induce degradation through ubiquitination. Instead, SUMO posttranscriptional modification (SUMOylation) of PDK4 was detected in MuRF1-overexpressing cells, in contrast to cells without the RING domain of MuRF1. MuRF1 deficiency enhances lipid metabolism possibly by upregulating PDK4 localization into mitochondrial through prevention of SUMOylation. Inhibition of MuRF1-mediated PDK4 SUMOylation is a potential therapeutic target for age-related dysfunction of lipid metabolism and muscle atrophy.
Abbreviations
-
- BW
-
- body weight
-
- COX IV
-
- mitochondrial cytochrome c oxidase subunit IV
-
- CSA
-
- cross-sectional area
-
- EDL
-
- extensor digitorum longus muscle
-
- EE
-
- energy expenditure
-
- GAPDH
-
- glyceraldehyde 3-phosphate dehydrogenase
-
- GA
-
- gastrocnemius muscle
-
- GFP
-
- green fluorescent protein
-
- HE
-
- hematoxylin-eosin
-
- MFN2
-
- mitofusin 2
-
- MM
-
- molecular mass
-
- MTS
-
- mitochondrial targeting signal
-
- MuRF1
-
- muscle RING finger 1
-
- MW
-
- muscle weight
-
- NEFA
-
- nonesterified fatty acid
-
- PDHs
-
- pyruvate dehydrogenase complex
-
- PDK4
-
- pyruvate dehydrogenase kinase 4
-
- PLA2
-
- phospholipase A2
-
- RQ
-
- respiratory quotient
-
- RT-qPCR
-
- quantitative reverse-transcription polymerase chain reaction
-
- SO
-
- soleus muscle
-
- SUMO
-
- small ubiquitin-related modifier
-
- TA
-
- tibialis anterior muscle
-
- ZT
-
- Zeitgeber time
1 INTRODUCTION
In advanced societies, age-related muscle atrophy, such as sarcopenia, is attracting attention as the target of research in this mechanism and becoming the therapeutic target against physical frailty.1, 2 Aging of skeletal muscle is characterized by atrophied muscle mass and a loss of force-generating capacity and leads to preferential loss of glycolytic and fast-twitch muscle fibers than oxidative and slow-twitch muscle.3, 4 Hence, age-related muscle atrophy affects the glycolytic function. On the contrary, many recent researches indicate that metabolic dysfunctions such as diabetes, hyperlipidemia, and obesity are causes of muscle atrophy.5, 6 Thus, muscle mass and metabolic function are closely related each other.
Protein degradation through ubiquitin-proteasome system along with the autophagy-lysosome system is the most important pathway associated with the mechanism of muscle atrophy. Muscle RING finger 1 (MuRF1) was first reported in 2001 as an unknown protein interacting with titin, which is located in M-Line of the myofibrillar sarcomere.7 Thereafter, MuRF1 was described as a striated muscle-specific ubiquitin ligase and a critical mediator of muscle atrophy caused by immobilization, denervation, and unloading.8 So far, many substrates of MuRF1 ubiquitination have been identified, including myosin heavy/light chains and alpha-actin.9, 10 At present MuRF1 is recognized as one of the most important atrophy-related genes, which are called “atrogenes”.11
Mice lacking MuRF1 are resistant to muscle atrophy caused by such conditions.8, 12, 13 Expression of MuRF1 is regulated at higher levels in glycolytic and fast-twitch muscle fibers than in oxidative and slow-twitch muscle fibers under the normal condition and during unloading or immobilization induced skeletal muscle atrophy.14 However, MuRF1 expression is reported to be increased, decreased, or hardly affected in skeletal muscles of aging rodents.15-17 Thus, the pathological role of MuRF1 in age-related muscle atrophy remains controversial.
Yeast two-hybrid screening showed that MuRF1 interacts with pyruvate dehydrogenase kinase 4 (PDK4), suggesting that PDK4 is a target substrate.18 PDK4 regulates energy production through a shift from glucose oxidation to fatty acid oxidation via pyruvate dehydrogenase (PDH) complex inhibition.19, 20 Cardiac-specific overexpression of PDK4 enhanced palmitate oxidation, but not glucose oxidation, thereby preventing diet-induced triglyceride accumulation in the heart.21 Interestingly, we found that PDK4 protein was accumulated into the mitochondria of MuRF1-deficient skeletal muscle, whereas MuRF1 failed to induce ubiquitination of PDK4. Recently MuRF1 reportedly interacted with small ubiquitin-related modifier-3 (SUMO3), a ubiquitin-like protein, involved in SUMO posttranscriptional modification (SUMOylation) via its RING domain.22, 23 Furthermore, PDK4 contains the conserved SUMOylation motif -ψ-K-X-E/D- (ψ: bulky hydrophobic residue), 282Gly-Lys-Glu-Asp, which binds SUMOs 1, 2, and 3.24
In this study, we elucidate pathological associations between MuRF1 and lipid metabolism in age-related atrophied muscles through interaction with PDK4 using MuRF1 knockout mice.
2 METHODS
2.1 Animals
MuRF1–/– mice, generated by inserting a neomycin-resistance gene into exon 2 of MuRF1, were kindly provided by Dr. Sorimachi.12, 25 MuRF1–/– mice were backcrossed with C57BL/6 mice more than eight times. MuRF1+/+ and MuRF1–/– male mice were housed in a room maintained at 23 ± 2°C on a 12:12-h light/dark cycle and allowed free access to standard chow and water. A small amount of blood for measuring glucose and insulin was collected after overnight fast 1 week before euthanasia. Body weight (BW) was measured monthly for 24 months. Fat mass of mice was quantified by computed tomography (LATheta LCT-200; Aloka Inc.).
Respiratory quotient (RQ) and energy expenditure (EE) were recorded every 10 min for 48 h using an Ox Imax™ sensor (Covidien-Nellcor). Mean values were calculated hourly, and these values were averaged at the same Zeitgeber time (ZT). Locomotor activity was measured using an ACTIMO system (Shintechno). The number of movements of mice recognized by infrared beams was counted for 48 h. Mean values were calculated at the same ZT.
All protocols were implemented according to the Guide for the Care and Use of Laboratory Animals at Tokushima University. Experimental protocols described in this study were approved by the Tokushima University Ethics Review Committee for Animal Experimentation.
2.2 Isolation of mitochondria
Mitochondria from COS7 and skeletal muscles from MuRF1+/+ and MuRF1–/– mice were prepared following previously established protocols.26 Briefly, cells or tissues were harvested and immediately minced in ice-cold CP-1 buffer (100 mM KCl, 50 mM Tris-HCl, 2 mM EGTA, pH 7.4). After grinding in a glass homogenizer, supernatants were passed through a 40-μm cell strainer. Mitochondria were then separated by differential centrifugation and lysed with RIPA™ buffer (50 mM Tris-HCl, pH 8.0, containing 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), 150 mM NaCl, 5 mM ethylenediaminetetraacetic acid [EDTA], and protease inhibitors) (Nacalai Tesque Inc.).
2.3 Plasmid constructs
Mouse MuRF1, PDK4, PDHα, PDHβ, and SUMO3 complementary DNAs (cDNAs) were isolated from a mouse skeletal muscle cDNA library by PCR. PCR products were subcloned into mammalian expression plasmid vectors pcDNA3.1-V5, pcDNA6.1-myc, and pcDNA-FLAG (MuRF1-V5, PDK4-myc, PDHα-myc, PDHβ-myc, and SUMO3-FLAG). RING domains of truncation mutants of MuRF1 (ΔRING-MuRF1-V5) and mitochondrial targeting signal (MTS) truncation mutants of PDK4 (ΔMTS-PDK4-myc) were constructed using a modified PCR technique (Stratagene Cloning Systems), as previously described.27 Construction of the expression plasmid for green fluorescence protein (GFP) containing the MTS of PDK4 (MTS-GFP) used an MTS mutant of PDK4 (residues 1–10, pcDNA6.1-MTS-myc) constructed as described above, and GFP was subcloned from the pCDNA3.1-GFP vector and ligated in-frame into the pcDNA6.1-MTS-myc vector.
2.4 Cell culture and transfection
COS7 cells were maintained in Dulbecco's modified Eagle medium containing 10% fetal bovine serum and penicillin-streptomycin (Nacalai Tesque Inc.) at 37°C in the presence of 5% CO2. Cells were transfected with plasmid vectors containing indicated genes using a FuGENE HD™ lipofection reagent (Roche Diagnostics) for 24 h before experiments.
2.5 SDS-polyacrylamide gel electrophoresis (PAGE) and Immunoblotting
Cells were homogenized with a sonicator in 50 mM Tris-HCl, pH 7.5, containing 150 nM NaCl, 1% Triton X-100, and protease inhibitors with EDTA. Protein samples were combined with 4× sample buffer (250 mM Tris-HCl, 8% SDS, 40% glycerol, 8% β-mercaptoethanol, 0.02% bromophenol blue) and separated on a polyacrylamide gel. Proteins were transferred to a polyvinylidene difluoride membrane and probed with the following primary antibodies, following the manufacturer's instructions: anti-V5 (R96025; Invitrogen), anti-myc (#2276; Upstate), anti-FLAG M2 (F3165; Sigma-Aldrich), anti-PDK4 (ab38242; Abcam), anti-PDH (ab110416; Abcam), anti-MFN2 (ab50838; Abcam), antimitochondrial cytochrome c oxidase subunit IV (COX IV) (#4844s; Upstate), anti-β-actin (A1978; Calbiochem), and anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (sc-25778; Santa Cruz Biotechnology). Secondary antibodies were donkey anti-rabbit 1:5000 and sheep anti-mouse 1:5000 (NA934-1ML, Amersham Biosciences and NA-9310, Piscataway). Membranes were developed using Amersham™ ECL™ or Amersham™ ECLTM Prime Western blotting detection reagents (GE Healthcare).
2.6 Immunoprecipitation
Immunoprecipitation samples were prepared using 100 μg protein and adjusted to 300 μl with lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, plus protease inhibitors with or without EDTA). Samples were then incubated with 0.5 μl of primary antibody at 4°C for 3 h under rotation. Thirty microliters of Protein G Sepharose™ 4 Fast Flow (GE Healthcare) was added to the samples, which were then rotated at 4°C for 16 h. The samples were washed in wash buffer six times and then prepared for SDS-PAGE as described above.
2.7 Quantitative reverse-transcription RT-qPCR
Total RNA was extracted from mouse skeletal muscle with an acid guanidinium thiocyanate-phenol-chloroform mixture (ISOGEN; Nippongene). RT-qPCR was performed using gene-specific primers and SYBR™ Green dye in an ABI StepOne™ PCR system (Applied Biosystems), as previously described.28 Oligonucleotide primers used for amplification are listed in Table 1.
| Target gene | Sequence | Length (bp) | |
|---|---|---|---|
| MuRF1 | S | 5′-ACGAGAAGAAGAGCGAGCTG-3′ | 179 |
| AS | 5′-CTTGGCACTTGAGAGGAAGG-3′ | ||
| PDK4 | S | 5′-AAAGGACAGGATGGAAGGAATCA-3′ | 82 |
| AS | 5′- TTTTCCTCTGGGTTTGCACAT-3′ | ||
| HSL | S | 5′-CCGCTGACTTCCTGCAAGAG-3′ | 213 |
| AS | 5′-CTGGGTCTATGGCGAATCGG-3′ | ||
| ATGL | S | 5′-GGTGACCATCTGCCTTCCAG-3′ | 193 |
| AS | 5′-TGCAGAAGAGACCCAGCAGT-3′ | ||
| PPARδ | S | 5′-GAGGGGTGCAAGGGCTTCTT-3′ | 101 |
| AS | 5′-CACTTGTTGCGGTTCTTCTTCTG-3′ | ||
| SREBP1c | S | 5′-GCAGACTCACTGCTGCTGAC-3′ | 150 |
| AS | 5′-AGGTACTGTGGCCAAGATGG-3′ | ||
| 18S ribosome | S | 5′-CATTCGAACGTCTGCCCTA-3′ | 137 |
| AS | 5′-CCTGCTGCCTTCCTTGGA-3′ | ||
| FAS | S | 5′-TGCTCCCAGCTGCAGGC-3′ | 107 |
| AS | 5′-GCCCGGTAGCTCTGGGTGTA-3′ | ||
| DGAT1 | S | 5′-GGAGACCGCGAGTTCTACAG-3′ | 183 |
| AS | 5′-CTCATGGAAGAAGGCTGAGG-3′ | ||
| SCD1 | S | 5′-ACCTGCCTCTTCGGGATTTT-3′ | 210 |
| AS | 5′-GTCGGCGTGTGTTTCTGAGA-3′ | ||
| CPT1b | S | 5′-CCCATGTGCTCCTACCAGAT-3′ | 130 |
| AS | 5′-CCTTGAAGAAGCGACCTTTG-3′ | ||
| ACC2 | S | 5′-GGCGATGAGCACCTTCTCTA-3′ | 61 |
| AS | 5′-CCCAGCCGAGTTTGTCACT-3′ | ||
| CD36 | S | 5′-GATGACGTGGCAAAGAACAG-3′ | 107 |
| AS | 5′-TCCTCGGGGTCCTGAGTTAT-3′ |
- Abbreviations: ACC2, acetyl-CoA carboxylase 2; AS, antisense primer; ATGL, adipose triglyceride lipase; CPT1b, carnitine palmitoyltransferase 00201b; DGAT1, diacylglycerol O-acyltransferase 1; FAS, fatty acid synthase; HSL, hormone-sensitive lipase; PPARδ, peroxisome proliferator-activated receptor δ; S, sense primer; SCD1, stearoyl-CoA desaturase-1; SREBP1c, sterol regulatory element binding protein 1c.
2.8 Measurement of the number of nuclei and fiber size of skeletal muscle
Sections of TA were stained with hematoxylin-eosin. Sections were photographed using a BIOREVO BZ-9000 fluorescence microscope (Keyence). The number of nuclei per muscle fiber and fiber size-frequency were measured in four sections of each muscle.
2.9 Measurement of biochemical parameters
Protein concentrations were determined using a bicinchoninic acid assay kit (Pierce).29 Serum concentrations of triglycerides and nonesterified fatty acid (NEFA) were measured using appropriate kits (Fuji Film Wako Pure Chemical Co.), following the manufacturer's protocol. Serum glucose concentrations were measured using a glucose meter for laboratory animals (Research and Innovation Japan Inc.). Serum insulin concentration was measured using an enzyme-linked immunosorbent assay kit (Funakoshi Co., Ltd.). Serum concentrations of pyruvate and lactate were determined using commercial assay kits (Funakoshi Co., Ltd.).
2.10 Cycloheximide (CHX) treatment
CHX is an inhibitor of protein synthesis, particularly of translation. CHX is widely used for evaluation of activity in protein degradation independent of protein synthesis. COS7 cells, transfected with the plasmid vectors containing the indicated genes, were treated with CHX for indicated periods.
2.11 Epoxomicin treatment
Epoxomicin is a strong and selective inhibitor of proteasomes and does not affect other protein degradation systems. COS7 cells, transfected with the plasmid vectors containing the indicated genes, were treated with epoxomicin for 2–4 h.
2.12 Statistical analysis
Data are expressed as mean ± SD (n = 3–7). Differences between two groups were assessed using Scheffé's test. All statistical analyses were conducted using SPSS 6.1 software (SPSS Japan). Differences were considered significant at p < .05.
3 RESULTS
3.1 Depletion of MuRF1 prevents body weight gain
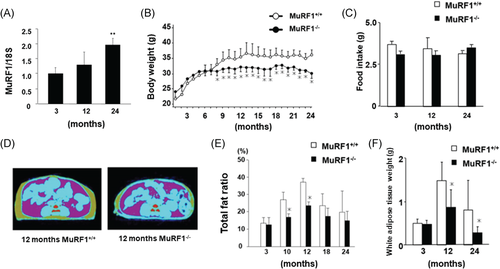
Gene expression of MuRF1 was measured in 3-, 12-, and 24-month-old mice in wild-type (MuRF1+/+) by RT-qPCR and increased with age (Figure 1A). Our RT-qPCR did not detect MuRF1 expression in any skeletal muscles of MuRF1-deficient (MuRF1–/–) mice (data not shown), indicating complete knockout of MuRF1 expression. We measured BW of MuRF1+/+ and MuRF1–/– mice monthly for 2 years. Interestingly, BW of MuRF1–/– mice more than 8 months old was significantly less than age-matched MuRF1+/+ mice (Figure 1B). However, no significant difference in food intake between MuRF1+/+ and MuRF1–/– mice was noted during the study period (Figure 1C). We assessed body composition using computed tomography scans to explore this finding in MuRF1–/– mice. The percentage of fat mass was significantly lower in MuRF1–/– mice than in MuRF1+/+ mice (Figure 1D,E). The wet weight of epididymal fat in 12- and 24-month-old MuRF1–/– mice was also reduced compared with MuRF1+/+ mice (Figure 1F), indicating that aged (>7 months old) MuRF1–/– mice were leaner.
3.2 MuRF1 deficiency partially protects from age-related muscle atrophy
We elucidated MuRF1 deficiency on age-dependent muscle atrophy, consistent with the association between muscle atrophy and ubiquitin ligase.8, 12, 13 We measured wet muscle weight of four hindlimb muscles (TA, extensor digitorum longus [EDL], gastrocnemius [GA], and soleus [SO]) at 3 and 24 months of age. Age-induced decreases in MW compensated by BW were significantly inhibited in TA and SO in MuRF1–/– mice (Figure 2A).
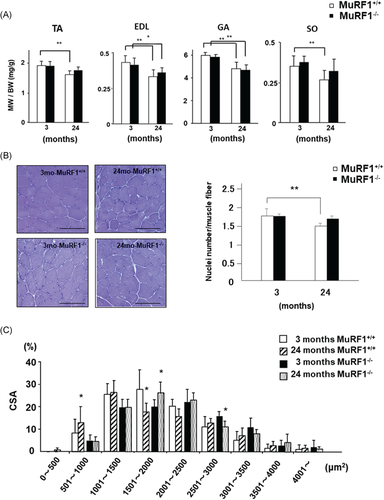
We next measured the number of nuclei per muscle fiber and cross-sectional area (CSA) of TA, which is sensitive to aging3, 4, 30 in mice at 3 and 24 months of age. The number of nuclei per muscle fiber significantly decreased in TA of MuRF1+/+ mice during aging. On the other hand, no significant change was observed in TA from MuRF1 deficient mice during aging (Figure 2B). In addition, the fiber size-frequency distribution of TA muscle fibers was measured. The fiber size-frequency distribution shifted to larger size in MuRF1–/– mice than in age-matched MuRF1+/+ mice (Figure 2C).
3.3 MuRF1–/– mice preferentially metabolize lipid rather than glucose
We measured serum and muscle (GA) parameters of lipid and glucose metabolism in 3 months old MuRF1+/+ and MuRF1–/– mice (Table 2) to determine why MuRF1 deficiency prevented BW and fat mass gain in mice. Already, at 3 months of age, the serum glucose levels in fasted MuRF1–/– mice were significantly lower than those in MuRF1+/+ mice, although the fasting serum insulin level of MuRF1+/+ mice was similar to that of MuRF1–/– mice. In contrast, serum NEFA levels in MuRF1–/– mice were significantly lower than that in MuRF1+/+ mice. Further, serum triglyceride levels tended to be lower in MuRF1–/– mice, although the difference with MuRF1+/+ mice was not significant. Interestingly, serum and muscle lactate levels were higher in MuRF1–/– mice than in MuRF1+/+ mice, but serum pyruvate levels were lower in MuRF1–/– mice. Thus, lipolysis and anaerobic glycolysis were promoted in the skeletal muscle of MuRF1–/– mice.
| MuRF1+/+ | MuRF1–/– | |
|---|---|---|
| Fasting glucose (mg/dl) | 92.7 ± 8.8 | 72.2 ± 9.1* |
| Fasting insulin (pmol/l) | 27.2 ± 16.9 | 29.9 ± 17.2 |
| NEFA (mmol/l) | 0.77 ± 0.10 | 0.53 ± 0.11* |
| Triglycerides (mmol/l) | 1.61 ± 0.31 | 1.40 ± 0.33 |
| Pyruvate (nmol/l) | 24.4 ± 4.5 | 18.9 ± 2.2* |
| Lactate (mmol/l) | 1.18 ± 0.07 | 1.21 ± 0.09* |
| Muscle lactate (nmol/mg protein) | 37.0 ± 1.5 | 62.3 ± 6.8* |
- Note: Biochemical parameters were measured in sera and gastrocnemius muscles of MuRF1+/+ and MuRF1+/+mice (3 months of age). Data are means ± SD (n = 5). *p < .05 versus MuRF1+/+ mice.
- Abbreviations: MuRF1, muscle RING finger 1; NEFA, nonesterified fatty acid.
We also analyzed whole-body lipid utilization and locomotor activity in young adult mice using indirect calorimetry and an animal movement analysis system, respectively. We use ZT, which shows time of lights on as ZT 0 and time of lights off as ZT 12. The RQ of 3-month-old MuRF1–/– mice was significantly lower than that of MuRF1+/+ mice (Figure 3A). In addition, EE of 3-month-old MuRF1–/– mice was significantly higher than that of MuRF1+/+ mice during ZT 0 to ZT 12 (light periods) in spite of similar locomotor activity. Even during ZT 12 to ZT 0 (dark periods), EE of MuRF1−/− animals was about the same as for MuRF1+/+ mice in spite of less locomotor activity (Figure 3A). These results indicate that MuRF1 deficiency enhanced EE, especially via lipid oxidation. Next, we measured gene expression of genes involved in lipogenesis and lipolysis and associated transcription factors in GAs of MuRF1+/+ and MuRF1–/– mice. MuRF1 deficiency showed little effect on expression of lipogenesis- and lipolysis-associated genes (Figure 3B).

3.4 PDK4 is accumulated in the mitochondria of MuRF1–/– skeletal muscle
A previous study showed that MuRF1 interacts with glucose and fatty acid metabolism-related molecules, such as PDHs and their negative regulators, PDKs, through yeast two-hybrid screens.18 We performed coimmunoprecipitation assays in MuRF1-V5 and empty (Mock)-, PDHα-, PDHβ-, or PDK4-myc cotransfected COS7 cells to confirm MuRF1 binding to these molecules. The assays revealed that PDK4 specifically interacts with MuRF1 (Figure 4A). We focused on PDK4 as a crucial enzyme for switching from glucose to fatty acid energy sources.19, 20
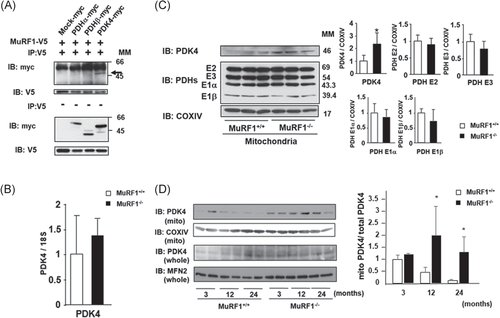
No significant difference in the gene expression of PDK4 was observed between the GA of MuRF1+/+ and MuRF1–/– mice (Figure 4B). Thus, we examined mitochondrial PDK4 protein levels in the skeletal muscle of MuRF1–/– mice; PDK4 translocated into the mitochondria after translation. Surprisingly, PDK4 significantly accumulated in the mitochondrial fraction of GA of 3-month-old MuRF1–/– mice (Figure 4C). In contrast, MuRF1 deficiency did not affect protein levels of PDHs, including PDH E1α, E1β, E2, and E3, in the mitochondrial fraction of GA (Figure 4C). Further, the increase of PDK4 protein levels of the muscle mitochondrial fraction in MuRF1–/– mice was sustained for 24 months. To confirm our hypothesis, we examined total PDK4 level by Western blotting. We used mitofusin 2 (MFN2), which is one of the mitochondrial fusion proteins, as internal standard, because its protein levels in human vastus lateralis muscle were not changed in a wide age range (21–88 years).31 We clearly demonstrated that the value of mitochondrial PDK4/total PDK4 in 12 and 24-month-old MuRF1–/– mice were higher than those in the respective month-old MuRF1+/+ mice. These results indicate that the accumulation of PDK4 was enhanced during aging (Figure 4D). These results suggest that MuRF1 affects translocation of PDK4 into skeletal muscle mitochondria in mice.
3.5 PDK4 interacts with MuRF1 possibly in cytosol
PDK4 is targeted to mitochondria via its N-terminal MTS (Met-Lys-Ala-Ala-Arg-Phe-Val-Met-Arg-Ser-).32 However, MuRF1 is in the cytosol.8, 12, 13 We thus examined the interaction of MuRF1 and PDK4 to explore mechanisms of interaction. We constructed an MTS-truncated mutant of PDK4 (ΔMTS-PDK4), which exhibits appropriate cytosolic localization, and a GFP containing the MTS of PDK4 (MTS-GFP), which exhibits appropriate mitochondrial localization (Figure 5A). We confirmed that PDK4 (-myc) localizes in the mitochondria; PDK4 was detected in the mitochondrial fraction containing MFN2, a mitochondrial protein. As expected, ΔMTS-PDK4 was detected in the cytosolic fraction containing cytosolic GAPDH (Figure 5B).
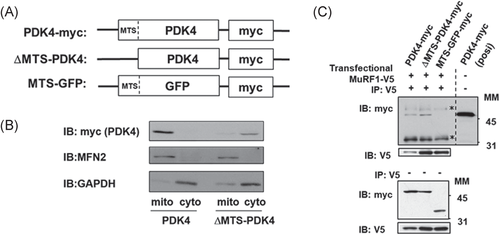
Coimmunoprecipitation assays revealed that MuRF1 interacted with ΔMTS-PDK4 and PDK4, but MTS-GFP failed to interact with MuRF1 (Figure 5C). Deletion of MTS in PDK4 enhanced interaction between MuRF1 and PDK4 (Figure 5C). Since MuRF1 interacted with native PDK4, which is preferentially localized in the mitochondria (Figure 5A), MuRF1 likely interacted with PDK4 during translocation from cytosol to mitochondria.
3.6 MuRF1 induces SUMOylation of PDK4, but not ubiquitination
MuRF1 is suggested to trigger muscle protein degradation through substrate ubiquitination.8, 12, 13 PDK4 accumulation in the mitochondria in MuRF1–/– skeletal muscle (Figure 4) led us to hypothesize a posttranslational modification by MuRF1. Therefore, we examined MuRF1 induction of ubiquitination and proteasomal degradation of PDK4. Cell lysates from cells transfected with mock vector or vectors containing MuRF1 or an inactive mutant with E2-binding domain deletion (ΔRING-MuRF1) were used in a PDK4 ubiquitination assay. Ubiquitination of PDK4 was measured in the presence of epoxomicin, a selective and strong proteasome inhibitor, to enhance the accumulation of ubiquitin conjugates. Overexpression of MuRF1 and ΔRING-MuRF1 failed to induce PDK4 ubiquitination (Figure 6A). Western blotting for PDK4 after CHX treatment showed that PDK4 degradation rate in MuRF1-expressing cells did not differ from degradation in mock- and ΔRING-MuRF1-expressing cells (Figure 6B). MuRF1 is probably not involved in ubiquitin-dependent degradation of PDK4.
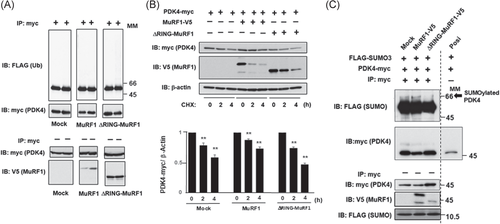
As described in introduction, MuRF1 reportedly interacts with SUMO3 involved in SUMOylation via its RING domain,22, 23 and PDK4 contains the conserved SUMOylation motif -ψ-K-X-E/D- (ψ: bulky hydrophobic residue), 282Gly-Lys-Glu-Asp, which binds SUMOs 1, 2, and 3.24 Thus, we examined whether MuRF1 induces SUMOylation of PDK4. The RING finger domain contains the SUMO2/3 binding site,23, 33 and ΔRING-MuRF1 was used as a negative control. Although SUMOylated PDK4 was weakly detected in the Mock and ΔRING-MuRF1 lane, we found that MuRF1 induced SUMOylation of PDK4 more than Mock and ΔRING-MuRF1 (Figure 6C).
4 DISCUSSION
MuRF1 is a typical E3 ubiquitin ligase associated with muscle atrophy and, along with muscle atrophy F-box protein 1/atrogin1,8 functions in the progression of muscle atrophy in various conditions. However, in age-related muscle atrophy, whether MuRF1 expression is increased, decreased, or unchanged as a function of age is controversial.15-17
In this study, we first examined the critical role of MuRF1 in age-related muscle loss using MuRF1-deficient mice. MuRF1 deficiency prevented age-related decrease in TA and SO weights, but not EDL and GA weights. Possibly, decrease in the gain of extra- and intra-muscular fat tissue during aging affected the wet MW of EDL and GA. Therefore, although MuRF1 deficiency prevented age-related decrease in the number of nuclei per muscle fiber and of CSA distribution in TA muscle, we suggest that MuRF1 deficiency partially prevents age-induced muscle atrophy. Although the data against muscle functions such as muscle force generation are lacking, much of the literature shows that MuRF1-deficient mice are resistant against muscle atrophy or muscle wasting. Since this study was designed to elucidate changes in lipid metabolism in MuRF1-deficient mice, there was no need for us to measure the muscle force generation.
However, these phenomena contradict a previous finding that muscle mass and fiber CSA are maintained with aging in MuRF1–/– mice, suggesting that MuRF1 deficiency prevents age-induced wet weight loss of all hindlimb skeletal muscles in mice.13 C/EBP-homologous protein, a maladaptive ER stress marker, was unchanged in the skeletal muscle of old MuRF1–/– mice. Authors concluded that with age, MuRF1 plays an important role in the control of skeletal muscle mass and growth capacity through the regulation of cell-level stress.13 In their study, MuRF1–/– mice were established by insertion of LacZ that simultaneously replaced approximately 8 kb of MuRF1 genomic sequences, comprising exons 1–4 and most of exon 5.13 In this study, murine MuRF1 was disrupted by homologous recombination into exon 2.12 This difference may explain the discrepancies between findings in the two studies.
Interestingly, MuRF1 deficiency prevented age-induced BW gain and adipose tissue accumulation. The results of indirect calorimetry and locomotor activity showed that MuRF1 deficiency enhanced the EE, especially through lipid metabolism. Significantly lower RQ and higher resting EE in MuRF1–/– mice are consistent with decreased serum levels of lipid metabolic parameters, such as NEFA and triglyceride. Prevention of BW gain by MuRF1 deficiency is thus possibly due to an increase in lipid metabolism. Obesity induces fat accumulation in both adipose and non-adipose tissues, such as the skeletal muscle and liver.34 Our findings in MuRF1–/– mice thus suggest a link between skeletal muscle and whole-body adipose tissue in the regulation of lipid metabolism. Although no past study has referred to the association between MuRF1 and fat tissue accumulation, a previous study proposed that the whole BW of an older MuRF1–/– mice represented an association.13 Our study showed significantly light BW of MuRF1–/– mice compared to that of MuRF1+/+ mice after 7 months of age. On the other hand, a previous study showed no difference in the BW between MuRF1+/+ and MuRF1–/– mice during aging. This difference might have originated from the deletion site of MuRF1, as described in the paragraph above. As a limitation, MuRF1 is a striated muscle-specific ubiquitin ligase.35, 36 Therefore, although a global deletion mouse model of MuRF1 has been accepted as a striated muscle-specific deleted model, there is some possibility of nonskeletal muscle effects, such as cardiac function, which was not evaluated in this study.
PDK4 is a crucial enzyme for the switch from glucose to fatty acid metabolism, and we focused on PDK4 function associated with MuRF1. Accumulated PDK4 was detected in skeletal muscle mitochondria of MuRF1–/– mice. In our study using COS7 cells with overexpression of MuRF1, MuRF1 likely interacted with PDK4 in cytosol and mediated SUMOylation but not ubiquitination of PDK4. This activity may inhibit mitochondrial translocation of precursor PDK4 protein in cytosol. We also considered that inhibition of PDH activity mediated by PDK4 accumulated in mitochondria stimulated mitochondrial lipid β-oxidation. This linkage is consistent with a previous finding that selective inhibition of PDHs by PDK4 resulted in enhanced lipolysis in the skeletal muscle during prolonged starvation.37
Cytosolic chaperones, such as heat shock protein 70 and mitochondrial import-stimulating factor, mediate import of proteins into mitochondria.38, 39 Protein modification in the cytosol is generally necessary for mitochondrial translocation. SUMOylation regulates protein stability, cytosolic-nuclear transport, and transcription.40 SUMOylation of some mitochondrial proteins such as dynamin-related protein 1 reportedly affects the localization and the function of the proteins.41, 42 PDK4 displays a consensus sequence for SUMOylation32; we detected SUMOylated but not ubiquitinated PDK4 in MuRF1-overexpressing cells. Deletion of MTS in PDK4 enhanced the interaction between MuRF1 and PDK4. We thus considered that SUMOylation mediated by MuRF1 is associated with localization and function of PDK4.
The effect of MuRF1 deficiency on the muscle weight is a local action, while PDK4-mediated metabolic changes on BW, total fat ratio, and lipid accumulation are systemic. The latter are preferentially dependent on volume, rather than kind, of skeletal muscles. Therefore, we used GA, a large muscle in the hindlimb, to elucidate changes in metabolic parameters, gene expression, and protein levels. Metabolic changes, including PDK4 SUMOylation in GA, are likely reflected in BW, total fat ratio, and lipid accumulation differences between MuRF1+/+ and MuRF1–/– mice. Also, previous literature suggests that MuRF1 suppresses glucose-related EE in the muscle by degradation of muscle creatine kinase.25 This outcome is consistent with our finding of decreased fasting serum glucose levels in 3-month-old MuRF1–/– mice. Thus, MuRF1 is also associated with regulation of lipid-related EE.
We used GA in 3-month-old young adult mice but not elder mice in the above analyses recognizing that changes in BW or adipose tissue accumulation in itself affects metabolism. We analyzed 3-month-old MuRF1–/– mice with BWs similar to MuRF1+/+ mice to eliminate the possibility that reduced BW and adipose tissue affect metabolism, independent of MuRF1 function. Also, PDK4 accumulation into mitochondria of MuRF1–/– mice is already notable at 3 months of age (Figure 4C). Occasionally, an obvious phenotype, such as obesity and leanness, requires several months after organ-level metabolic changes, although the mechanism of such a time lag is unknown.
Differences in food intake between MuRF1+/+ and MuRF1–/– mice might influence BW during aging, even if such differences are not significant. However, gain in BWs in MuRF1–/– mice fed with a high-fat diet for 10 weeks was smaller than MuRF1+/+ mice fed the same diet, although their food intakes were similar (data not shown). Therefore, it seems unlikely that differences in daily food intake caused differences in body weight between MuRF1+/+ and MuRF1–/– mice.
We first detected impaired body weight gain in striated muscle-specific MuRF1-deficient mice during aging. Therefore, we investigated the effect of increased MuRF1 during aging by using MuRF1-deficient mice. An in vivo experiment using MuRF1-deficient mice was important for evaluating local reaction, such as the phenotype of skeletal muscle as well as systemic reaction, such as BW, accumulation of fat tissue, and metabolic function like RQ or EE against the deletion of MuRF1. It is exceedingly difficult to observe such a linkage only via an in vitro model. Thus, we suggested the possibility of the effect of MuRF1 against lipid metabolism via PDK4 localization. To investigate this association between the presence of MuRF1 and PDK4 localization in detail, we performed an in vitro experiment using transfection of the plasmid vector with mutant ΔMTS-PDK4 and ΔRING-MuRF1. From these experiments, we suggested the possibility that MuRF1 suppressed lipid metabolism by inhibiting PDK4 transfer into the mitochondria, possibly through posttranslational SUMOylation of PDK4. Similarly, we considered that the MuRF1 knockout model is especially useful for researching the inter-organ linkage associated with changes in the muscle phenotype.
A part of our study used in vitro experiments with COS7 cells overexpressing MuRF1, because we could not adequately transfect the plasmid vector into differentiated C2C12 myotubes. In addition, when we transfected the plasmid vector with Mock, MuRF1, and ΔRING-MuRF1 into the cells. Cells that did not express MuRF1 originally were of interest in this study. Because non-SUMOylated PDK4 and changes in PDH activity and phosphorylation were not detected in skeletal muscles of MuRF1–/– mice, could possibly indicate a smaller ratio of SUMOylated PDK4 to non-SUMOylated PDK4. Further studies are necessary to elucidate these issues.
In this study, we newly discovered that increasing MuRF1 expression during aging is associated with muscle mass decrease through ubiquitin-proteasome system as well as lipid metabolism dysfunction through the regulation of localization of PDK4 protein. Muscle mass maintenance is important against the homeostasis of energy metabolism, and the converse is also true. Hence, MuRF1 might be a key protein that regulates age-related sarcopenia and metabolic diseases. In the near future, inhibitors or repressors of MuRF1 might be developed as a therapeutic target against locomotive and metabolic dysfunction during aging.
5 CONCLUSION
MuRF1 deficiency partially prevented age-related weight loss of skeletal muscle. However, BW and body fat tissue of more than 7-month-old MuRF1–/– mice were lower than those of age-matched MuRF1+/+ mice, indicating that aged MuRF1–/– mice were leaner. The lower RQ and higher EE of 3-month-old MuRF1–/– mice suggest that MuRF1–/– mice preferentially metabolize lipid as the energy source. PDK4 was accumulated in the skeletal muscle mitochondria of 3-month-old MuRF1–/– mice compared with age-matched MuRF1+/+ mice. In vitro study using MuRF1 overexpressing COS7 cells, MuRF1 interacted with PDK4 in cytosol, and mediated SUMOylation of PDK4. Therefore, we suggest that MuRF1 deficiency enhances lipid metabolism by upregulating localization of PDK4 into mitochondria through the inhibition of SUMOylation. Inhibition of MuRF1-mediated PDK4 SUMOylation is a potential therapeutic target to the age-related dysfunction of lipid metabolism and obesity in addition to age-related muscle atrophy.
ACKNOWLEDGMENTS
We thank the late Dr Sorimachi at Tokyo Metropolitan Institute of Medical Science and the late Dr. Edward M. Mills the Division of Pharmacology and Toxicology, Faculty of Pharmacology, Texas University for their helpful suggestions and providing knockout mice. We would like to thank Enago (https://www.enago.jp/) for English language editing. This work was supported by Grant-in-Aid for Scientific Research (KAKENHI) (Grant Number: 18H04981 and 19H04054) from JSPS, and AMED-CREST (Grant Number: 19gm0810009h0104), Japan.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Conceptualization: Kosuke Sugiura, Katsuya Hirasaka, and Takeshi Nikawa; Data curation: Kosuke Sugiura, Katsuya Hirasaka, Tasuku Maeda, Takayuki Uchida, Koji Kishimoto, Motoko Oarada, and Anayt Ulla; Formal analysis: Katsuya Hirasaka; Funding acquisition: Takeshi Nikawa; Methodology: Kosuke Sugiura, Katsuya Hirasaka, Motoko Oarada, Iori SakakibaraReiko Nakao, Siegfried Labeit; Project administration: Takeshi Nikawa; Validation: Katsuya Hirasaka, and Koichi Sairyo; Writing—original draft: Kosuke Sugiura and Katsuya Hirasaka; Writing—review and editing: Siegfried Labeit, and Takeshi Nikawa. All authors have read and agreed to the published version of the manuscript.




