Inhibition by EGTA of the formation of a biofilm by clinical strains of Staphylococcus aureus
Abstract
The effect of EGTA on the adhesion and on the formation of a biofilm by two reference and eight clinical strains of Staphylococcus aureus was studied. All the clinical strains were isolated from patients from Kinshasa. Spa typing confirmed that these clinical strains were distinct. The Biofilm Ring Test (BFRT®) showed that EGTA (100 µM–10 mM) inhibited the adhesion of the four clinical methicillin-resistant (MRSA) strains and the crystal violet staining method that it inhibited the formation of a biofilm by all the strains. Divalent cations abolished the effect of EGTA on the formation of a biofilm, specially in the clinical MRSA strains. EGTA had no effect on established biofilms. Only concentrations of EGTA higher than 10 mM were toxic to eukaryotic cells. Our results establish the effectiveness and the safety of lock solutions with EGTA to prevent the formation in vitro of biofilms by S. aureus.
Abbreviations
-
- BFI
-
- Biofilm Index
-
- BHI
-
- brain heart infusion
-
- BFRT®
-
- Biofilm Ring Test®
-
- CVSM
-
- crystal violet staining method
-
- IBS
-
- initial bacterial suspension
-
- PIA
-
- polysaccharide intercellular adhesin
Introduction
Staphylococcus aureus is a Gram positive bacterium which is one of the leading causes of nosocomial infections. It is for instance responsible for half of the infections of prosthetic devices 1. This high prevalence of S. aureus in infections of medical material can be explained by the high propensity of the bacteria to form a biofilm 2. Biofilms have been defined as colonies of bacteria which adhere to each other or to a surface and which develop within an organic matrix 3. This mode of growth is the usual way of bacterial life. The ability of S. aureus to form a biofilm on biotic as well as on abiotic surfaces probably explains why it is responsible for persistent infections 4.
Biofilm formation by S. aureus is secondary to the expression of the polysaccharide intercellular adhesin (PIA), which mediates intercellular adhesion 5. PIA is a polymer formed of N-acetylglucosamine and is synthesized by proteins encoded by the genes of the intercellular adhesion locus ica 6, 7. But strains carrying deletions of this operon can also form a biofilm demonstrating the existence of an alternative pathway 8. Whatever the mechanism, it remains that the formation of the biofilm proceeds in a well-established sequence: the primary adherence of the bacteria to a surface is followed by the accumulation of interacting bacteria forming a multilayered biofilm 9. The dispersion of the bacteria occurs when the biofilm has reached some maturity. The matrix forming the core of the biofilm mainly contains polysaccharides, proteins and DNA. Within the biofilm, bacteria gain resistance to antimicrobials 10 and against host defences 11. The chronicity of the infections involving biofilms is probably secondary to the occurrence of persisters 12. We have recently reported that the adhesion of S. aureus on an abiotic surface can be explored with the Biofilm Ring Test (BFRT®) 13 performed as described by Chavant et al. 14. Using this method, we reported that both MRSA and MSSA strains adhered to an abiotic surface within few (2–3) h. This method was rapidly saturated and could not be used to determine the growth of the biofilm with time. The crystal violet staining method (CVSM) was much more useful to measure the biofilm formed after 1 or 2 days. Using this second method, we could demonstrate heterogeneity among various clinical strains of S. aureus, some of them forming a very important biofilm after 24 h 13. The aim of the present work was to study the sensitivity of various strains of S. aureus to EGTA, a chelator of divalent cations (mostly magnesium, calcium and iron) 15. We tested the effect of the chelator on the initial adhesion of bacteria, on their capacity to develop a strong biofilm or on the destruction of a preformed biofilm.
Materials and methods
Origin of the S. aureus strains
The eight clinical strains used in this work were collected for diagnostic purposes in the Laboratory of Bacteriology of the Provincial General Reference Hospital of Kinshasa (HGPRK) in the Democratic Republic of Congo (Table 1). One strain (the 027/U strains) was isolated from urine, 1 strain (the 011/LP strain) from prostatic fluid, two strains (the 007/FV and the 028/FV strains) from smear tests, two strains (the 5668/B and 5741/B strains) from blood samples, and two strains (the 1532/SW and 1620/SW strains) from skin wounds. The two reference strains (the MSSA ATCC-25923 and the MRSA ATCC-33591 strains) tested in this study for comparison with clinical strains were obtained from the American Type Culture Collection (Manassas, VA, USA). The bacteria were grown and isolated on Brain Heart Agar with 5% v/v sheep blood and on Mannitol Salt Agar (Difco, BD Franklin Lakes, NJ, USA). The identification of S. aureus was performed with the latex agglutination test (Pastorex Staph-Plus, BioRad, Marnes-la-Coquette, France). Antibiograms of each isolated strain were realized using the diffusion method on Mueller Hinton agar with the following antibiotic disks (Liofilchen, Roseto degli Abbruzzi, Italy): amikacin, amoxicillin, cefazolin, ceftazidime, ciprofloxacin, gentamicin, oxacillin, and vancomycin. The sensitivity of the strains to methicillin was examined with the diffusion method on the Mueller Hinton agar with 4% NaCl. The results of the susceptibility tests were analyzed as reported in a previous publication 13. Resistance to methicillin for S. aureus was also determined with the cefoxitin disk diffusion method.
| Genes | Primers | Size of the amplicon (bp) | |
|---|---|---|---|
| mecA | F | 5′-AAAATCGATGGTAAAGGTTGGC | 533 |
| R | 5′-AGTTCTGCAGTACCGGATTTGC | ||
| Nuc | F | 5′-GCGATTGATGGTGATACGGTT | 279 |
| R | 5′-AGCCAAGCCTTGACGAACTAAAGC | ||
| 16 S rRNA | F | 5′-GTTATTAGGGAAGAACATATGTG | 750 |
| R | 5′-CCACCTTCCTCCGGTTTGTCACC |
Genomic characterization of the Staphylococcus aureus strains
DNA extraction
DNA was extracted from the bacteria as described by Unal et al. 16. Briefly, isolates cultured for 24 h on Columbia agar with 5% sheep blood were successively incubated with lysostaphin and proteinase K, boiled, and finally centrifuged. The lysate obtained was used as the template in multiplex PCR and spa typing.
Multiplex assay for the detection of 16S rRNA, mecA and nuc genes
The 16S rRNA, mecA and nuc genes were detected as described by Maes et al. 17. The multiplex PCR assay was performed with 50 µl of reaction mixture containing 1× PCR buffer, 2 mM MgCl2, 16S rRNA-specific primers (0.6 µM) and mecA and nuc-specific primers (0.4 µM each), 250 µM of each of the four dNTPs, 2 U Taq DNA polymerase and 5 µl of DNA lysate. The primers were supplied by Amersham Pharmacia, Biotek (Roosendaal, The Netherlands) (Table 1). The PCR conditions were modified as follows: denaturation at 95 °C for 15 min, followed by 25 cycles of denaturation at 95 °C for 30 s, annealing at 57 °C for 90 s, extension at 72 °C for 90 s, and a final extension at 72 °C for 10 min.
spa typing
The spa typing was based on the method described by Hallin et al. 18. The polymorphic X region of the protein A gene (spa) was amplified from isolates by using the forward primer: 5′-TAAAGACGATCCTTCGGTGAGC-3′ and the reverse primer: 5′-CAGCAGTAGTG CCGTTTGCTT-3′ 19. The sequences of the amplicons were determined with the ABI Prism BigDye Terminator v1.1 Sequencing Kit (Applied Biosystems, Darmstadt, Germany). The nucleotide sequences were analyzed using the RIDOM Staph-Type software (Ridom GmbH, Germany) to assign the isolates to the various spa types.
Measure of the growth rate of S. aureus
S. aureus strains were grown in brain heart infusion (BHI) medium to stationary phase with constant shaking (100 rpm) at 37 °C. Two-hundred and fifty microliters cells in a stationary phase were inoculated to 50 ml fresh BHI medium and 200 µl of this bacterial suspension were transferred in a 96-wells plate. The plates were incubated at 35 °C under constant shaking in a microplate reader (Synergy HT, BioTek, Winnoski VT, USA). The wells were read every 5 min for 6 h at 600 nm. The data collected were fitted to an exponential equation and the doubling time was calculated using the GraphPad software. For each analysis the coefficient of correlation of the fitting was higher than 0.98.
Evaluation of the biofilm formation with the BFRT®
The formation of the biofilm was evaluated using the BFRT® kit (BioFilm Control, Saint-Beauzire, France) as previously described 20. The assays were performed in sterile 96-wells polystyrene plates formed by the assembly of 12 individual eight-well strips (StripWell MSW002B). After extensive homogenization for 1 min, a solution of very small (1–3 µm) positively charged magnetic beads (TON005N) was added to the Initial Bacterial Suspension (IBS) containing ∼107 cells ml−1 (assays) or to BHI medium (controls) to obtain a final concentration of 10 µl of magnetic beads ml−1. The wells were inoculated with 200 µl of the IBS-Toner mixture (magnetic beads). Each strain was studied in triplicate. Two wells served as the control wells and were inoculated with the BHI-Toner mixture. The plates were covered with a lid; they were transferred in a humid atmosphere and incubated at 35 °C. At the end of different incubation times, 100 µl of contrast liquid (inert opaque oil) provided with the kit were added to each well. This inert opaque oil facilitated the reading of the plate by the dedicated Scan Plate Reader (BioFilm Control). The wells were read before and after 1 min exposure to a magnet included in the Block Test (BioFilm Control). The images were analyzed with the software provided by BioFilm Control. The adhesion of the bacteria on the bottom of each well was estimated using the Biofilm Index (BFI) based on the size of the black spot formed by the attracted beads and detected at the bottom of the wells. The calculated BFI for a well was inversely proportional to the number of adherent cells in this well. The variation of the BFI (ΔBFI) for a well was calculated by subtracting the BFI of the tested well (BFIsample) from the mean of the BFI of the two control wells (BFIcontrol). The experiment was repeated at least three times, for each strain and incubation time.
Evaluation of the formation of a biofilm with the CVSM
Polystyrene sterile strips were inoculated with 200 µl of IBS and incubated for various times at 35 °C in a humid atmosphere. A control well was inoculated with sterile medium. Each strain was evaluated in triplicate. The medium was removed from the wells, which were washed three times with 200 µl sterile distilled water. The strips were air-dried for 45 min and the adherent cells were stained with 200 µl of 0.1% Crystal violet solution. After 45 min, the dye was removed and the wells were washed five times with 300 µl of sterile distilled water to remove excess stain. The dye incorporated by the cells forming a biofilm was dissolved with 200 µl of 33% v/v glacial acetic acid and the absorbance of each well was read at 540 nm in the microplate reader. The results were expressed as the variation of OD540 nm (OD540 nm sample − OD540 nm control). The experiment was repeated at least three times, for each strain and incubation time.
Evaluation of the toxicity of EGTA on eukaryotic cells
The effect of EGTA on the viability of eukaryotic cells was assessed by measuring the ability of human monocytes/macrophages (cell line THP-1) to reduce 3-(4,5-dimethylthiazole-2-yl)-2,5-diphenyl tetrazolium bromide (MTT). This reaction is catalyzed by oxydoreductases, which use electrons from mitochondria-generated NADH as the primary reductants 21. The cells were grown at 37 °C in 5% CO2 atmosphere in RPMI 1640 medium supplemented with 10% heat inactivated fetal calf serum, 2 mM L-glutamine, 100 U ml−1 penicillin, 100 µg ml−1 streptomycin (Gibco, Groningen, The Netherlands). When confluent, the cells were centrifuged and seeded in 96-well plates at 100,000 cells well−1 (total volume 200 µl well−1), in the presence of 100 nM Phorbol Myristate Acetate (PMA) (Sigma–Aldrich, St. Louis MO) in order to differentiate them into mature macrophage-like cells 22. Eighteen hours after seeding, the cells were washed once with culture medium and 200 µl of medium with the various concentrations of EGTA was added to each well. The cells were incubated for 2 h at 37 °C, the medium was removed before the addition to the wells of 100 µl MTT solution in Hanks Balanced Buffer Solution (1 mg ml−1 medium). After 3 h incubation of the cells with MTT, formazan crystals develop in living and early apoptotic cells. These crystals were then solubilized in dimethylsulfoxide and the absorbance was measured at 540 nm in the microplate reader. Each condition was tested in sextuplicates.
Detection of selected genes coding for surface proteins involved in the formation of a biofilm
DNA was extracted from one or two colonies of each strain. The cells were suspended in 20 ml sterile distilled water and heated at 100 °C for 20 min in a water bath. The detection of icaA, femB, and mecA genes has been described previously 13. The primers used for the amplification of the new genes studied in this work were listed in Table 2. These primers were supplied by Eurogentec (Liège, Belgium). Five microliters of the bacterial DNA solution were added to 45 µl of PCR mixture containing 16 mM ammonium sulfate, 67 mM Tris–HCl (pH 8.8), 200 µM of each of the four dNTPs, 2 mM MgCl2, 2 µM of each sasG primer, 0.5 µM of each sasC, fnbA, or fnbB primers, 0.4 µM of each clfA or clfB primers and 1.25 U Taq DNA polymerase. The primers used in this study were specifically designed for S. aureus (for references see Table 2). The sequences were amplified using a Bio-Rad icycler according to the protocols listed in Table 2. A hot-start PCR protocol was used and the samples were incubated for 3 min at 94 °C before running the PCR protocols. After 30 cycles, the samples were incubated for 10 min at 72 °C. Five microliters of the various PCR media were analyzed by a 1% (clfA and gap genes) and 2% (all the other genes) agarose gel electrophoresis, the gels were stained with Gel Red (VWR, Leuven, Belgium) and visualized using a UV light in a Chemidoc XRS+ apparatus (BioRad, Nazareth, Belgium). The size of the amplified fragments was estimated by comparison with the GeneRuler 100-bp plus DNA ladder (Fermentas, Hanover, MD, USA).
| Gene | GenBank | Primers | PCR Conditions | Size (bp) | References | |
|---|---|---|---|---|---|---|
| sasC | 2859120 | F | 5′-GCAACGAATCAAGCATTGG- | 94 °C (60 s); 50 °C (60 s); 72 °C (60 s) | 489 | 23 |
| R | 5′-AGACAGCACTTCGTTAGG- | |||||
| sasG | 11932242 | F | 5′-GGGAACTCAACAAGAGGCAG- | 94 °C (60 s); 50 °C (60 s); 72 °C (60 s) | 311 | 24 |
| R | 5′-CAGAACGAGCTTTTCTAACC- | |||||
| fnbA | 3913735 | F | 5′-GATACAAACCCAGGTGGTGG- | 94 °C (30 s); 55 °C (30 s); 72 °C (30 s) | 191 | 25 |
| R | 5′-TGTGCTTGACCATGCTCTTC- | |||||
| fnbB | 3238062 | F | 5′-GGAGCGGCCTCAGTATTCTT- | 94 °C (30 s); 55 °C (30 s); 72 °C (30 s) | 201 | 25 |
| R | 5′-AGTTGATGTCGCGCTGTATG- | |||||
| clfA | 5330438 | F | 5′-CCGGATCCGTAGCTGCAGATGCACC- | 94 °C (60 s); 55 °C (60 s); 72 °C (60 s) | 1000 | 26 |
| R | 5′-GCTCTAGATCACTCATCAGGTTGTTCAGG- | |||||
| clfB | 5331889 | F | 5′-ACATCAGTAATAGTAGGGGGCAAC | 94 °C (60 s); 55 °C (60 s); 72 °C (60 s) | 205 | 27 |
| R | 5′-TTCGCACTGTTTGTGTTTGCAC | |||||
| Gap | 1123535 | F | 5′-ATGGTTTTGGTAGAATTGGTCGTTTA | 94 °C (60 s); 50 °C (60 s); 72 °C (60 s) | 934 | 28 |
| R | 5′-GACATTTCGTTATCATACCA | |||||
| mupR | X75439.1 | F | 5′-TATATTATGCGATGGAAGGTTGG | 94 °C (60 s); 50 °C (60 s); 72 °C (60 s) | 458 | 29 |
| R | 5′-AATAAAATCAGCTGGAAAGTGTTG |
- DNA from each strain was extracted and the detection of the various genes was performed by PCR as described in Materials and Methods Section. The size of the amplicons was calculated by applying the program GeneRunner on the sequences of the genes found in the GenBank.
Statistical analysis
The statistical analysis was performed with GraphPad Prism 4.0. Results were analyzed using a two-way ANOVA followed by a Bonferroni post-test performed for each dose. *** p < 0.005; ** p < 0.01; * p < 0.05.
Results
Characterization of the clinical MSSA and MRSA strains
The five MRSA strains and the reference MSSA strain have already been characterized 13. In this previous study, we showed that three MRSA clinical strains were resistant and that the 028/FV strain was sensitive to cefoxitin. But the five MRSA strains harbored the mecA gene. The four new clinical MSSA strains, the 1532/SW, 1620/SW, 5668/B, and 5741/B strains were first characterized (data not shown). These four strains were sensitive to methicillin. The triplex PCR results showed that all strains (MSSA and MRSA) were positive for 16S rRNA and nuc genes. The typing of the strains used in the present study was performed by analyzing the spa gene. As shown in Table 3, seven spa types were identified in this study; one MSSA strain (1532/SW) was spa nontypeable. This result established that the eight clinical strains tested in this work had different genetic backgrounds.
| MRSA | mecA | nuc | 16 S rRNA | spa type | Cefoxitin |
|---|---|---|---|---|---|
| Strains | |||||
| 007/FV | + | + | + | t002 | R |
| 011/LP | + | + | + | t044 | R |
| 027/U | + | + | + | t051 | R |
| 028/FV | + | + | + | t2265 | S |
| MSSA | |||||
| 5668/B | − | + | + | t355 | S |
| 5741/B | − | + | + | t939 | S |
| 1532/SW | − | + | + | Non typeable | – |
| 1620/SW | − | + | + | t10715 | S |
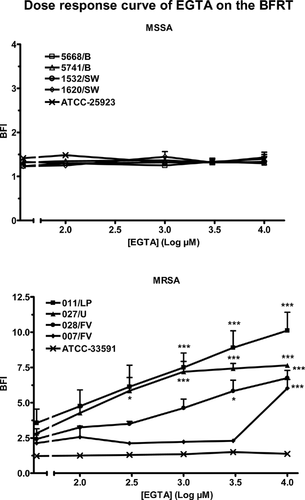
Effect of EGTA on the immobilization of micro-magnetic beads in the BFRT®
Control experiments (incubations of the magnetic beads in the presence of 10 mM EGTA, in the absence of bacteria) established that the ion chelator had no effect on the mobility of the beads in a magnetic field (data not shown). The effect of EGTA on the immobilization of the beads by the eight clinical strains and the two reference strains was tested during 6 h experiments. The sensitivity to EGTA varied among the 10 strains. The five MSSA (clinical and reference) strains were totally insensitive to concentrations of EGTA in the 100 µM to 10 mM range (upper panel, Fig. 1). The immobilization of the beads by the reference MRSA strain was also not affected by the presence of EGTA in the medium. The four clinical MRSA strains were inhibited by EGTA but not with the same sensitivity (lower panel, Fig. 1). One millimolar EGTA blocked the immobilization of the beads by the 011/LP strain (BFI from 3.6 ± 0.9 to 7.5 ± 1.0), the 027/U strain (BFI from 2.8 ± 0.3 to 7.2 ± 0.7) and the 028/FV strain (BFI from 2.4 ± 0.1 to 4.6 ± 0.6). The fourth clinical MRSA strain (007/FV) was not affected by 1 mM EGTA (BFI from 2.1 ± 0.6 to 2.2 ± 0.1); it was inhibited by a ten-fold higher concentration of EGTA (increase of the BFI to 6.1 ± 0.6).
Effect of EGTA on the formation of the biofilm measured with the CVSM
The biomass (bacteria plus extracellular polymeric matrix) adhering to the bottom of the wells after 6 h was estimated with the CVSM. As shown in Fig. 2, the absorbance measured in the wells after incubation in control conditions averaged 0.328 ± 0.028 (n = 52) for MSSA strains and 0.160 ± 0.007 (n = 36) for MRSA strains (p < 0.001). Adding EGTA to the culture medium dose-dependently decreased the absorbance measured with all the strains. The EC50 of EGTA averaged 48 µM for the five MSSA strains and 26 µM for the four clinical MRSA strains. The difference between the MSSA and MRSA strains was not significant. The interaction between cations and EGTA was tested on biofilms formed during 24 h. As shown in Table 4, calcium, magnesium, and manganese cancelled the inhibitory effect of EGTA in MRSA strains. In MSSA strains, only manganese could significantly abolish the effect of EGTA.

| MRSA strains | MSSA strains | |||
|---|---|---|---|---|
| OD540 nm | N | OD540 nm | n | |
| CONT | 0.124 ± 0.003 | 12 | 0.530 ± 0.040 | 12 |
| EGTA | 0.076 ± 0.004* | 8 | 0.278 ± 0.013* | 12 |
| +Ca | 0.139 ± 0.005** | 7 | 0.340 ± 0.020 | 11 |
| +Mg | 0.137 ± 0.003** | 8 | 0.353 ± 0.013 | 11 |
| +Mn | 0.150 ± 0.006** | 8 | 0.447 ± 0.025** | 12 |
- A biofilm was formed for 24 h in control conditions (CONT) or in the presence of 1 mM EGTA alone (EGTA) or EGTA with 1 mM calcium, magnesium or manganese. The biofilm was assayed with the CVSM. Results are expressed as the OD540 nm and are the means ± SEM of n experiments.
- * p < 0.005 when compared to incubation in control conditions.
- ** p < 0.005 when compared to incubation in the presence of EGTA and in the absence of added cation.
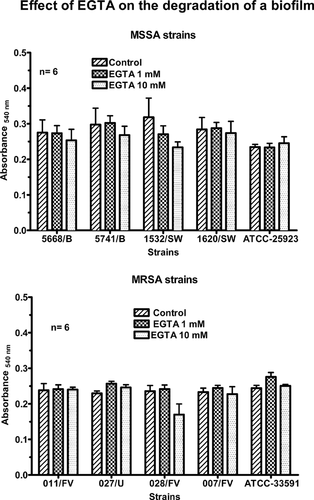
Effect of EGTA on a preformed biofilm
The previous results confirmed that EGTA not only inhibited the initial adhesion of some strains but also prevented the subsequent formation of the biofilm by all the strains. In the next experiment, the ability of EGTA to trigger the destruction of a biofilm was tested. The biofilm was formed during the culture of the bacteria in 96-wells plates for 24 h. After removal of the culture medium, the bacteria were further incubated for 4 h in control conditions or in the presence of 1 or 10 mM EGTA. The biomass present in the wells was then estimated by the CVSM. As shown in Fig. 3, the chelator had no significant effect on the absorbance of Crystal violet in any strain. These results suggested that EGTA did not destroy preformed biofilms.
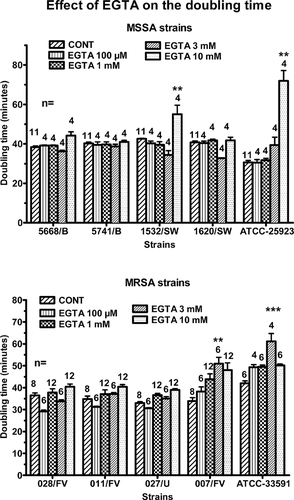
Effect of EGTA on the growth of the bacteria
The inhibition by EGTA of biofilm formation might be secondary to an inhibition of the growth of the bacteria. To test this hypothesis, planktonic cultures were performed either in control conditions or in the presence of increasing concentrations of EGTA and the doubling times of the various strains were measured (Fig. 4). The doubling time of the MSSA strains averaged 39 ± 1 min. EGTA had no effect except at a 10 mM concentration, which increased by 25% the doubling time of the 1532/SW strain and by 120% the doubling time of the reference strain. The doubling time of the MRSA strains averaged 36 ± 1 min. EGTA had only slight effects. The most significant effects were observed at a 3 mM concentration, which increased by 50% the doubling time of the 007/FV strain and by 30% the doubling time of the reference strain.
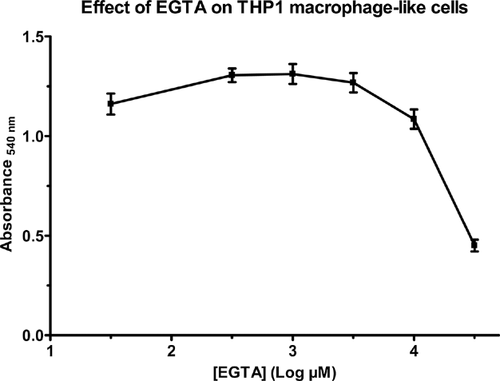
Toxicity of EGTA towards eukaryotic cells
The toxicity of EGTA on THP1 macrophage-like cells was tested with the MTT test. This test explores the ability of the cells to transfer electrons and to reduce a dye. Human macrophages were incubated for 2 h with various concentrations of EGTA. As shown in Fig. 5, the chelator had no effect on the absorbance at concentrations ranging from 1 to 10 mM. At 30 mM, the highest concentration tested, EGTA decreased the absorbance (from 1.085 at 10 mM EGTA to 0.451 at 30 mM EGTA). These results showed that low concentrations of EGTA had no toxic effects on the viability of the human cells.
Detection of the genes coding for some surface proteins
In a last experiment, the genes coding for proteins involved in antibiotic resistance and for surface proteins contributing to bacterial adhesion were searched by PCR amplification. All the MRSA strains were mecA+ and femB+. The 007/FV strain was the only MRSA strain, which was sasG− and mupR+. All the MSSA strains were mecA− and femB+. They were also sasC− and sasG− except for the 5741/B strain (sasC+ and sasG+) and the 1620/SW strain (sasC+). All the MRSA and the MSSA strains were fnbA+, fnbB+, clfA+, clfB+, icaA+, and gap+.
Discussion
In the present study, we showed that, at low concentrations, which were non-toxic to eukaryotic cells, EGTA prevented the adhesion and/or the development of a biofilm by clinical strains of S. aureus, but had no effect on established biofilms. Our results were fully consistent with those of Shanks et al. 30 who reported that citrate, which binds extracellular calcium, inhibited the formation of a biofilm by S. aureus, but was unable to destroy preformed biofilm. Oulahal et al. 31 observed that EGTA increased the effect of ultrasound on the removal from stainless steel of E. coli milk but not of S. aureus milk biofilms.
The dynamics of biofilm formation has been extensively studied and two major steps have been clearly distinguished 9. Initially, S. aureus adheres on a biotic (e.g. apical membrane of epithelial cells) or on an abiotic surface (e.g. contact lenses). We explored this initial adhesion with the Biofilm Ring Test® 20. After 4 h, all the strains nearly fully immobilized the magnetic beads and the BFI decreased from values ∼17 to values ∼1.5 (MSSA strains) or ∼2.5 (MRSA strains). At that time, the strains formed a biofilm, which could be stained with Crystal violet. The adhesion on an abiotic surface (BFRT®) and the formation of a biofilm (CVSM) were not significantly different between the MSSA and the MRSA strains.
The adhesion and formation of a biofilm by the 10 strains were differently affected by the presence of EGTA in the medium. At a concentration of 1 mM, the chelator had no effect on the adhesion of all the MSSA strains and of the reference and of one clinical (007/FV) MRSA strains but it significantly inhibited the adhesion of the 3 other clinical MRSA strains (BFRT®). It decreased the formation of a biofilm by all the staphylococcal (MRSA and MSSA) strains (CVSM). At this concentration, EGTA had no effect on the doubling time of all the strains confirming that these inhibitions were not related to a lower number of bacteria. The divergent results obtained with EGTA on adhesion and formation of the biofilm confirmed that these two properties are distinct and are differently regulated by cations, presumably calcium. Gristina et al. 32 reported that, in the presence of magnesium and calcium, extracellular polysaccharides polymerize and form a gel. The cations promote the adhesion of bacteria on surfaces. Dunne and Burd 33 also observed that divalent cations increased in vitro and under static conditions, the adhesion of S. epidermidis to plastic. Calcium, magnesium and manganese reversed the inhibition of EGTA on the formation of a biofilm by MRSA strains. Only manganese was effective on MSSA strains. The affinity of manganese for EGTA (pKa 12.2) is better than calcium (pKa 10.9) and much better than magnesium (pKa 5.3). As a result, the concentration of free EGTA should be lower in the presence of 1 mM Mn than in the presence of the two other cations 34. According to Taweechaisupapong and Doyle 35, chelating agents are inhibitors of several bacterial adhesins. Our results showing different sensitivity of MRSA and MSSA strains to EGTA also suggest that some clinical strains not only acquire resistance to antimicrobials but also modify their surface properties. Furthermore, Jonsson and Wadström 36 showed that clinical strains of S. aureus might express hydrophilic capsule polymers contributing to their invasive capacity and to their resistance to phagocytosis. This was illustrated by the results of Tachi and Hirabayashi 37. Using a rat model of skin wound, they reported that a clinical strain of S. aureus penetrated deeper in the wound than a reference strain. Molina-Manso et al. 38 demonstrated that a collection strain and clinical strains of S. aureus differently adhered on ultra-high molecular weight polyethylene treated with vitamin E.
More recently, Abraham et al. 39 reported that the effect of citrate and EGTA on biofilm formation varied among strains. They inhibited the formation of a biofilm in strains like the Newman strain and increased the formation of the biofilm in the related 10833 strain. The deletion of the gene coding for the clumping factor B abolished the biofilm-forming property of the two calcium chelators. In an attempt to determine which genes might account for the different sensitivity of adhesion to EGTA, the genes coding for major surface proteins involved in adhesion or formation of a biofilm were studied by PCR. The results of these PCR showed very subtle differences among the 10 strains. The gene sasC could be detected in all strains except the 5668/B and 1532/SW MSSA strains. The protein SasC is a surface protein mediating cell aggregation and biofilm accumulation 23. The absence of amplicon for sasG and the presence of an amplicon for mupR were the only differences between the 007/FV strain and the other MRSA strains. The protein SasG is a surface protein contributing to the adherence and the formation of a biofilm by S. aureus 40. It has also been reported that the strains expressing this protein could form a biofilm even in the absence of the PIA 40. None of the differences among the amplified genes could explain the different behavior of the strains in the absence of calcium. The genotyping of MRSA and MSSA clinical isolates by spa typing method revealed seven different spa types (four spa types for MRSA and three types for MSSA strains). The MSSA strain 1532/P was non-typeable due probably to the lack of spa gene. Among the MSSA strains, the spa type t355 has been isolated in clinical strains from some African countries such as Mali 41, Gabon 42, 43, and Nigeria 44. The spa type t939 has been recently identified in isolates from Gabon 42, 43. In MRSA clinical strains, the spa type t002 is found in isolates from Europe 45 and from Central and Western African countries 42-44. The spa types t044 and t051 are found in many European countries 45, but the spa type t044 was recently identified in strains from Tunisia 46. Our results suggest intercontinental exchanges of several clones. Travelers are now considered as an important source of dissemination of S. aureus infections around the world. The analysis of the spa gene also revealed the existence of a new subtype (t10715). This is the first report of this subtype in a clinical MSSA strain not only from African isolate but also on a world-wide scale. This type (15-12-17-17-16-02-16-02-25-17) belongs to the spa clonal complex 012 (15-12-16-02-25-17-24-24) and has probably arisen after successive insertions, deletions and duplications leading to the subtypes t021 (15-12-16-02-25-17-24), t421 (15-12-16-02-25-17), t342 (15-12-16-02-16-02-25-17) and t1848 (15-12-17-16-02-16-02-25-17).
In conclusion, the Biofilm Ring Test® and the Crystal violet staining technique used in this study have nicely contributed to demonstrate that low concentrations of EGTA, non-toxic to eukaryotic cells, prevent the adhesion of some bacterial cells to an abiotic surface and inhibit the development of the biomass of the biofilm. Our results also show that, once established, the biofilm becomes resistant to EGTA, suggesting that such EGTA-containing lock solutions should be used to prevent the formation of biofilms but not to promote their removal.
Acknowledgments
This work was supported by grant no. 3.4577.10 of the Fonds National de la Recherche Scientifique of Belgium (F.N.R.S.) and by a grant from the Coopération Technique Belge (C.T.B.). J.M. Liesse Iyamba is a recipient of a Research Fellowship from the C.T.B. and C. Nagant is a Research Associate of the F.N.R.S. The authors thank Pr M. Vandenbranden (Faculty of Sciences, Free University of Brussels, Belgium) for his generous gift of THP-1 cells.




