Genotypic intraspecies heterogeneity of Enterococcus italicus: data from dairy environments
Abstract
The diversity of a collection of 19 Enterococcus italicus strains isolated from different dairy sources was explored using a molecular polyphasic approach, comprising random amplification of polymorphic DNA (RAPD-PCR), repetitive element PCR (REP-PCR), plasmid profiling and ribotyping. The data obtained showed a high-level of biodiversity, not always correlated to the niche of isolation. Particularly, REP-PCR with primer BOXA1R and plasmid profiling allowed the best discrimination at strain level. Exploiting the genome shotgun sequence of the type strain of the species, available in public database, genes related to insertion sequences present on enterococcal Pathogenic Islands (ISEf1, IS905), determinants related to virulence factors (codifying for hemolysin and cell wall surface proteins), exogenously DNA (conjugal transfer protein, replication plasmid protein, pheromone shutdown protein, phage integrase/recombinase) and penicillin binding proteins system were detected. The presence of most of these genes seemed a common genetic trait in the Enterococcus genus, sur gene (cell wall surface protein) was only detected in strains of E. italicus. To our knowledge, this is the first time that specific primers, with the expection of the species-specific probe targeted to 16S rRNA gene, have been designed for this species.
Introduction
Over the past decade, the sequences of more than 100 bacterial genomes have become available in the public domain. Considerable attention has focused on pathogenic bacteria, including food-borne pathogens. This progress includes elucidation of the partial genome sequences of more than 20 lactic acid bacteria 1. The genus Enterococcus encompasses a considerable number of different species (http://www.bacterio.net) that display a relatively large degree of diversity 2. However, the only complete and fully annotated Enterococcus genome sequence accessible and published to date is that of Enterococcus faecalis V583 3. For E. faecium, E. gallinarum and E. casseliflavus whole genome shotgun sequences are deposited. The availability of these genetic information's and post-genomic tools opens new avenues to investigate the intraspecies variability that could provide a deeper insight into the metabolic interactions of enterococci during food processing, as well as their role in pathogenesis.
Recently, within the frame of the Human Microbiome Project 4 the genome of E. italicus DSM15952T, the type strain of the species, has been sequenced and deposited in a public database (Accession: PRJNA61487, ID: 61487).
The species E. italicus has been proposed a few years ago for a set of seven strains isolated from cows' milk used in the production of artisanal Italian cheeses 5, 6. Since that time, other strains have been isolated and characterized from cheeses of different geographical areas 7-9. In previous studies 10 we showed that E. italicus possess a poor acidifying activity, as well as low proteolytic and autolytic properties, in agreement with data obtained for other enterococcal species 11. A further study on the potential virulence profiles 10 underlined a different behavior of the dairy strains (low antibiotic resistance rates and the absence of resistance and virulence genes) in comparison with the other enterococcal clinical strains 12, 13. All E. italicus strains tested neither use transposition nor posses transmissible or mobilizable plasmids 14. However, the identification of a clinical isolate as E.italicus 15, and the finding of a strain containing a transmissible plasmid 16 indicate the need to improve knowledge of this bacterial species.
It is known that the degree of bacterial biodiversity is an important factor influencing both positive and negative properties of the species. For this reason, in this work we studied the level of intraspecies variability in E. italicus species.
Since the number of available E. italicus strains for a detailed study is still limited, we focused firstly on the isolation of novel strains of the species. For that purpose we also screened ecological niches so far not investigated. Based upon the strains isolated, we applied a molecular polyphasic approach using random amplification of polymorphic DNA (RAPD-PCR), repetitive element PCR (REP-PCR), plasmid profiling, ribotyping and genome analysis. RAPD-PCR is a rapid molecular genotyping tool to study the biodiversity, successfully applied on lactic acid bacteria 17, 18. REP-PCR represents an easy to perform fingerprinting method for grouping and tentatively identification of microorganisms, based on primers complementary to certain repetitive sequences dispersed in bacterial genome 19. These methods are frequently used in bacterial taxonomy and they have been successfully applied for reliable and fast differentiation at species level of different bacterial groups including lactobacilli 20 and enterococci 21. Ribotyping is another PCR method based upon differences in rRNA, which generates a highly reproducible and precise fingerprint (ribotype) that can be used to classify bacteria from the genus through and beyond the species level. In bacterial genomes the ribosomal operon (rrn) is composed principally of housekeeping genes. The genetic variation in these genes is primarily responsible for ribotype polymorphisms 22. Species genotypically homogeneous are characterized by one or only a few ribotypes, while heterogeneous species show polymorph restriction patterns 23. Since, mobile genetic elements (MGE) are responsible for genome plasticity and intraspecies varability 24 the presence of MGE has been investigated as well in this study. Horizontal transmission of genes and MGE, are a sources of intraspecies variation 25. Enterococci are prone to acquire antimicrobial resistance either by point mutation or by horizontal transfer of MGE 26. Finally we explored the shotgun genome sequence of the type strain of E. italicus deposited in Gene Bank to investigate the intraspecies variation based on the genetic content 27.
Materials and methods
Sample material and bacteriological analysis
Fifty six samples of commercially available cheeses from different region of Italy, four from Lithuania and other two respectively from Perù and Mexico, made with ewe, goat or cow milk were analyzed. At the same time ten samples from cow, pork, poultry meat, two samples from human and cat excrements respectively, and one from olives and their brine were analyzed. The presence of E. italicus was verified in cheese samples directly by total DNA extraction. For the other matrices a conventional enrichment step in M17 broth (Difco Laboratoires, Detroit, USA) supplemented with 1% (w/v) glucose (M17g) was firstly carried out (incubation at 37 °C). Samples (10 g) tested positive were homogenized with 10 volumes of a sterile saline (0.9% NaCl, w/v) solution in a laboratory paddle blender (Stomacher 400, Seward, London, UK). Decimal dilutions were prepared in saline solution and plated in duplicates for viable counts on M17g agar. After 24 h of incubation at 37 °C colony numbers was calculated, characteristic colonies were randomly selected and subcultivated (purified) on M17g plates.
Bacterial strains and growth conditions
Enterococcus italicus strains included in this study and reference strains used for comparison are listed in Table 1 All strains were maintained as frozen stocks at −80 °C using glycerol as cryoprotective agent; reactivation of strains was done in M17g at 37 °C overnight.
| Strain | Source | Reference |
|---|---|---|
| E. italicus TP1.5T (DSM 15952T) | Toma piemontese cheese | 5 |
| TP1.3 | Toma piemontese cheese | 5 |
| TP2.3 | Toma piemontese cheese | 5 |
| TP1.D | Toma piemontese cheese | 5 |
| TP3.D | Toma piemontese cheese | 5 |
| RP1 | Robiola piemontese cheese | 5 |
| RP4 | Robiola piemontese cheese | 5 |
| EIP1 | Pannerone cheese | this study |
| EIP2 | Pannerone cheese | this study |
| EIP3 | Pannerone cheese | this study |
| EIP4 | Pannerone cheese | this study |
| EIP5 | Pannerone cheese | this study |
| EIP6 | Pannerone cheese | this study |
| EIP7 | Pannerone cheese | this study |
| EIP8 | Pannerone cheese | this study |
| EIP9 | Pannerone cheese | this study |
| EIP10 | Pannerone cheese | this study |
| ITAC1 | Mexican cheddar cheese | this study |
| ESAa | Belgian cheese | 7 |
| E. saccharolyticus DSMb 20726T | Straw bedding | 38 |
| E. sulfureus DSM 20481T | Plant material | 39 |
| E. faecium ATCCc 19434T | Clinical speciment | 40 |
| E. faecalis ATCC 19433T | Clinical speciment | 40 |
| E. faecalisV583 ATCC 700802 | Human blood | 41 |
| E. avium DSM 20679T | Human faeces | 42 |
| E. gallinarum DSM 20628T | Chicken intestine | 42 |
- a Type strain of Enterococcus saccharominimus LMG 21727T, later synonym of Enterococcus italicus 43, LMG: Belgian Co-ordinated Collection of Microorganisms-Bacterial Collection, Gent, Belgium.
- b DSM: German Collection of Microorganism and Cell Cultures, Braunschweig, Germany.
- c ATCC: American Type Culture Collection, Rockville, Maryland, USA.
DNA extraction
Total DNA of each strain was extracted as described previously 28, plasmidic DNA was isolated by the alkaline extraction procedure as described by Anderson and McKay 29. Further purification of DNA was performed according to published procedures 30. Genomic DNA for all PCR reactions was extracted from a 100 μl aliquot of an overnight culture diluted with 300 μl TE 1 × buffer (10 mM Tris-HCl, 1 mM Na2EDTA, pH 8.0) as described by Mora et al. 31.
Ribotyping
The genomic DNA of Enterococcus italicus isolates (ca. 10 μg) was digested by incubation with 30 U of HpaII endonuclease according to manufacturer's instructions (MBI-Fermentas, Vilnius, Lithuania). A 20 μl aliquot of the digestion mixture was combined with 5 μl of loading buffer and was electrophoresed on a 1.2% (w/v) agarose gel at 100 V for 2 h. Subsequently, DNA fragments were transferred to a nylon membrane (Roche Diagnostics GmbH, Mannheim, Germany) by Southern Blot, according to the manufacturer's instructions 30. Hybridization was performed at 60 °C using E. italicus DSM 15952T16S rDNA. The probe was produced with primers and conditions previously reported 32. For digoxigenin labeling of the 1500 bp fragment the DIG DNA Labeling and Detection kit (Roche) was used. Pre-hybridization and hybridization were performed in 50% (w/v) formamide at 42 °C overnight and stringency washes in 0.1 × SSC buffer at 65 °C (10 SSC buffer consisted of 1.5 M NaCl and 150 mM sodium citrate). Digoxigenin-labelled probes that hybridized to a target sequence were detected with a digoxigenin luminescence detection kit (Roche) using CSPD {3-/4-methoxyspiro[1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.1(3,7)decan]-4-yl) phenyl phosphate; signals were recorded on X-ray film for 2 h.
Genotypic analysis
Isolates were grouped by combined analysis of repetitive element (REP) typing using primer BOXA1R (5′- CTACGGCAAGGCGACGCTGACG-3′) and (GTG)5 (5′-GTGGTGGTGGTGGTG-3′) (PRIMM, Milan, Italy) as reported by Colombo et al. 33 and Versalovic et al. 34 respectively, and random amplification of polymorphic DNA-PCR (RAPD) typing using primers M13 (5′-GAGGGTGGCGGTTCT-′3) as described by Rossetti and Giraffa 35.
Data analysis
The experiments were repeated a minimum of three times to confirm the band patterns and only consistent and reproducible band patterns were used for data analysis. The gels were marked for the presence (1) or absence (0) of the corresponding band in the different isolates. The binary data generated was analyzed for genetic similarity using unweighted pair group arithmetic mean (UPGMA) program of NTSYSpc software, version 2.11 36.
Primers
The identification of potentially new E. italicus strains was conducted with species-specific primers targeted to 16S rRNA gene 37. Primers used for the detection of specific genes coding for ISEf1 (IS905-like transposase), IS256, traK (conjugal transfer protein, NZ_AEPV01000090), traB (pheromone shutdown protein, NZ_AEPV01000079), repE (replication plasmid protein, NZ_AEPV01000111), int (phage integrase/recombinase, NZ_AEPV01000033), sur (cell wall surface protein, NZ_AEPV01000004), hem (hemolysin, NZ_AEPV01000066), mrcB (penicillin-binding protein 1B, NZ_AEPV01000037), pbpB (penicillin-binding protein 2B, NZ_AEPV01000041), pbp1A (penicillin-binding protein 1A, NZ_AEPV01000044), pbp2A (peni-cillin-binding protein 2A, NZ_AEPV01000087) and pbpC (penicillin-binding protein C, NZ_AEPV01000003) were designed on the basis of the E. italicus DSM15952T whole genome shotgun sequence (Accession: PRJNA61487, ID: 61487). The sequences of primers are specified in Table 2.
| Gene(s) | Primer pair (5′-3′) | Amplicon (bp) |
|---|---|---|
| ISEf1 | F: CCTAGAGATCGGAATGG | 727 |
| R: GGCAAATAATAAAAAGTTAA | ||
| IS256 | F: AGCGAAGAGATTCAAACG | 995 |
| R: TTCAATTAGATTGGTACT | ||
| traK | F: ACTTTCTTGCGACGTGTCGA | 518 |
| R: TCGTTGCTTGTGCTGCCTTT | ||
| traB | F: GCGAACCTCGACGAGAACAA | 422 |
| R: AAGGTGAAGAGTGCCGCAAG | ||
| repE | F: CAAGTCGACGAACCCTGGTT | 364 |
| R: ACTTGCGAACAGCTCGCTC | ||
| int | F: GCGCTCGTTCCTTTGAACTG | 953 |
| R: ATCCGAGTGGGTCAAGGCTT | ||
| sur | F: ACAACAGCTGGTGAGGGAGT | 805 |
| R: TGGTCGTTGTAGCCCCATCA | ||
| hem | F: CTGTCGGCTTCGACAAGCTT | 891 |
| R: TGGCTCGTCATCCTCGAAGT | ||
| mrcB | F: ATTGGTACCGAGACACGGGT | 652 |
| R: ACGCTGTTCCATCGTCATGG | ||
| pbpB | F: TGCAGAGCATGCCTACGAAC | 540 |
| R: ACCACCGCAGCCTGAAATAC | ||
| pbp1A | F: GGAAGCACCTTGACACAGCA | 961 |
| R: CGACACCCACATCTGCGAAT | ||
| pbp2A | F: GTGACAGCCTTAGTCGGTCG | 387 |
| R: CAGCGGTGACTCGCCATATT | ||
| pbpC | F: CCATTGCTGAGGATGCGACT | 520 |
| R: TCTTTGGCAGCCACGGATTC | ||
| ita | F: TACCGCATAATACTTTTTCTCT | 323 |
| R: GTCAAGGGATGAACATTCTCT |
DNA amplification procedure and PCR conditions
Each 25 μl reaction mixture contained 100 ng of bacterial DNA, 2.5 μl of 10 × reaction buffer, 200 μM deoxynucleoside triphosphate, 2.5 mM MgCl2, 0.5 μM of each primer and 0.5 U of Taq polymerase (MBI-Fermentas). The amplification was performed in a Gene Amp PCR System 2400 (PerkinElmer, Norwalk, CT). PCR conditions: initial denaturation step (2 min 94 °C) was followed by 35 cycles of denaturation at 94 °C for 1 min and annealing at 55 °C for 1 min, with extension at 72 °C for 2 min. The final cycle was followed by an additional 7 min elongation period at 72 °C. PCR products were separated on 1.5% (w/v) agarose gel stained with ethidium bromide in 1 X Tris-acetate-EDTA buffer (40 mM Tris-acetate and 1 mM EDTA, pH 8.0) and photographed under UV light.
Results
Isolation and distribution of E. italicus in sample matrices
We analyzed different food matrices, out of 74 samples, for the presence of E. italicus, with particular regards to cheese (62 samples), meat (10), olives and brine (2) and human (1) and cat excrements (1). After species-specific PCR, the expected amplicon of 323 bp was obtained only from total DNA of dairy samples (10%) suggest-ing that dairy habitat may be one favourite ecosystem of E. italicus. Twenty-five randomly selected colonies grown on M17g agar plates for each PCR positive dairy sample were collected. In total, 150 novel isolates were subjected for identification by molecular analyses using E. italicus species-specific probe. Only from Pannerone and Mexican cheddar cheese E. italicus was isolated (Table 1). From other cheese samples, despite the positive signal in PCR using species-specific probe, E. italicus strains were not obtained by cultivation. The distribution of E. italicus is most likely related to the type of cheeses and/or stage of ripeness. In Pannerone and Toma-like cheeses E.italicus seems to be a typical member of the bacterial population, while in other kind of cheeses this species may be only sporadically present.
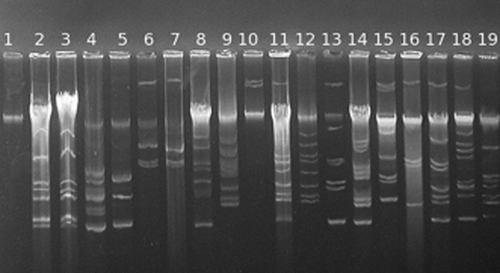
Plasmid profiling
In all E. italicus isolates we detected bands indicative for plasmids, only the type strain (DSM 15952T) tested negative. The isolates contained two to nine plasmids, some with high molecular mass (Fig. 1). Two E. italicus strains (TP1.3 and TP2.3) had the same plasmid profile although, as reported in Fortina et al. 10, they are different isolates.
Molecular typing
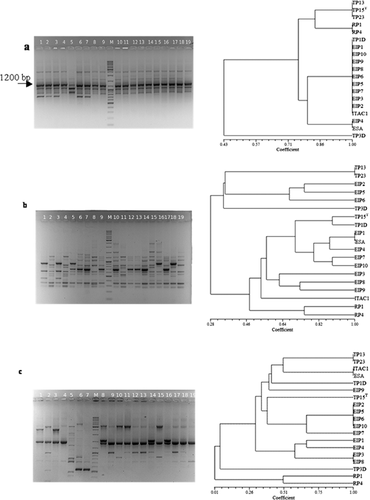
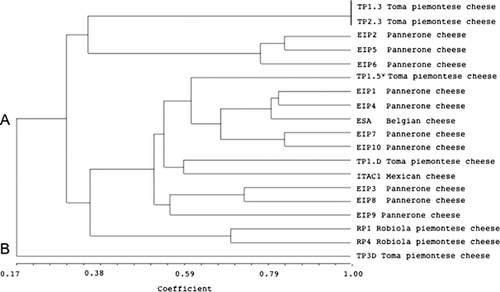
Three different primer sets (Fig. 2) were used for the evaluation of E. italicus genetic fingerprinting. Primer (GTG)5 showed low discriminatory ability (Fig. 2), only five electrophoretic profiles were obtained. However, the presence of a common band of about 1200 bp in all strain tested was interesting. More heterogeneous patterns were obtained with the primers BOXA1R and M13 (Fig. 2). Dendrograms obtained from UPGMA analyses demonstrated that these primers are different in their discriminatory power. BOXA1R primer clustered 16 groups (and/or strains), primer M13 grouped 19 strains in 13 clusters. The isolates obtained from Robiola cheese and isolate TP3.D from Toma piemontese cheese were separated from the other isolates at similarity level ranging from 1 to 30% using M13 primer (Fig. 2). Within the main cluster it was possible to observe different level of correlation among the strains. Some isolates showed a unique band profile, whereas other strains (EIP2, 5, 6, 10 Pannerone cheese) showed a band pattern similarity of 100%. REP-PCR using primer BOXA1R allowed a better discrimination at strain level, 16 of the 19 strains showed a unique fingerprint (Fig. 2). UPGMA dendrogram obtained combining M13 and BOXA1R profiles (Fig. 3) further indicated a high-level of biodiversity, represented by two different strain clusters. Cluster B contained the unique isolate TP3.D and cluster A was split into different sub-clusters. For cluster A no correlation between ecological niche and isolates could be observed.
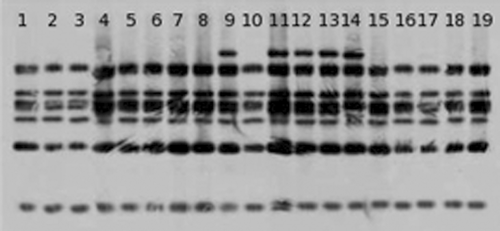
Ribotyping
Two different clusters of E. italicus strains with seven or eight bands in their riboprints could be identified by PCR-ribotyping (Fig. 4). One cluster was characterized by seven bands, including all strains isolated from Toma piemontese and Robiola cheese as well as isolates from Pannerone cheese (EIP1, EIP3, EIP9 and EIP10), Mexican cheddar (ITAC1) and Belgian cheese (ESA). The ribotype fingerprints of the other isolates were characterized by eight bands and formed separate cluster.
Genome analysis
Virulence factor typing showed that all 13 genes screened are detectable in E. italicus strains (Table 3). Whereas, the reference strains of the other tested species gave variable results. Genes for phage integrase/recombinase (int), hemolysin (hem) and the penicillin binding protein (pbpB) were detected in all enterococci.
| Strain/gene | E. italicus TP1.5 DSM 15952T | TP1.3 | TP2.3 | TP1.D | TP3.D | RP1 | RP4 | EIP1 | EIP2 | EIP3 | EIP4 | EIP5 | EIP6 | EIP7 | EIP8 | EIP9 | EIP10 | ITAC1 | ESAa | E.saccharolyticus DSMb 20726T | E.sulfureus DSM 20481T | E.faecium ATCCc 19434T | E.faecalis ATCC 19433T | E.faecalisV583 ATCC 700802 | E.avium DSM 20679T | E.gallinarum DSM 20628T |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ISEf1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | – | – | + | + | – | – |
| IS256 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | – | – | – | + | – | – |
| traB | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | – | – | + | – | – |
| traK | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | – | – | + | – | – |
| int | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| sur | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | – | – | – | – | – | – | – |
| repE | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | – | + | + | – | + | – | – |
| hem | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| mrcb | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | – | – |
| pbpB | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| pbp1A | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | – | + | + | – | – |
| pbp2A | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | – | + | + | – | – |
| pbpC | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | – | + | + | – | – |
- a Type strain of Enterococcus saccharominimus LMG 21727T, later synonym of Enterococcus italicus 43, LMG: Belgian Co-ordinated Collection of Microorganisms-Bacterial Collection, Gent, Belgium.
- b DSM: German Collection of Microorganism and Cell Cultures, Braunschweig, Germany.
- c ATCC: American Type Culture Collection, Rockville, Maryland, USA.
Gene for surface cell wall protein (sur) was only detected in strains of E. italicus.
Discussion
The genus Enterococcus is one of the most controversial group of lactic acid bacteria. Studies on the microbiota of many traditional cheeses in the Mediterranean countries have indicated that enterococci play an important role in cheese ripening. Enterococcus spp. are also present in other fermented foods, such as raw sausages and olives. At the same time, enterococci have also been associated with a raising number of human infections. The aim of the polyphasic study presented here was (1) the isolation of a broader number of E. italicus strains and (2) the molecular genetic analysis of the biodiversity of this species.
Identification at intraspecies level by molecular typing is an important issue to determine the identity and relatedness of industrial or clinical relevant bacterial strains. For RAPD analysis we used primer M13, and for REP-PCR we used BOXA1R and (GTG)5 primers. Overall, the primer (GTG)5 determined a lower number of amplicons than the primers M13 and BOXA1R (Fig. 2), but highlighting the presence of common band of 1200 bp that could be a nucleotide region characterizing this new species. BOXA1R and M13 primers generated complex restriction patterns and were more suitable for strain differentiation (Fig. 2). Also using ribotyping analysis it is possible to evidentiate some interesting differences, whose significance remain still unclear.
Although it was not possible to relate the high degree of biodiversity found to the dairy ecosystem source, the presence of specific strain genotypes emphasizes the importance of raw milk as a source of strain genetic diversity in the cheeses. Plasmid profiling showed that E. italicus isolates have distinct profiles, this underlines the genome heterogeneity of the species. It seems that plasmid profiling is a useful technique for the differentiation of E. italicus strains.
In a previously study 10 we screened for virulence determinants such hemolysin, integrase and surface wall protein, but using primers designed on E. faecalis, E. italicus tested negative. In this study we used specific primers designed on genome shotgun sequence data of E. italicus DSM 15952T. The presence of genes encoding for phage integrase/recombinase (int), hemolysin (hem) and penicillin binding protein (pbpB) in all tested may be a common genetic trait in the genus, sur gene (cell wall surface protein) was only detected in E. italicus strains. This gene sequence could be used for an E. italicus species-specific molecular probe.
Today, the availability of the complete nucleotide sequences of several bacterial genomes represents a further opportunity for the study of gene expression profile. The presence of the shotgun sequence of E. italicus DSM 15952T is an important point of departure for studying the biology of the species. To our knowledge, this is the first time that specific primers, with the exception of the species-specific probe targeted to 16S rRNA gene 37, have been created for E. italicus (sur gene). Moreover, new primer sets for insertion sequences, ISEf1 and IS256, related to enterococcal Pathogenic Islands, have been designed.
The combined use of RAPD, REP-PCR, ribotyping and plasmid profiling is a good strategy to characterize novel isolates of dairy origin and to verify species heterogeneity of Enterococcus spp. Research will continue on the expression of detection virulence genes in E. italicus.
Acknowledgements
This work was supported by “Post genomica batterica per la qualità e sicurezza degli alimenti” project from the Lombardy region (Italy).




