Two separate domains in the golli myelin basic proteins are responsible for nuclear targeting and process extension in transfected cells
Abstract
The golli products of the myelin basic protein (MBP) gene are expressed in neurons and oligodendrocytes (OLs). In certain neuronal populations, golli proteins undergo translocation between the nucleus and cytoplasm/processes during development. The proteins consist of two domains, a golli domain of 133 amino acids and an MBP domain of variable length. One objective of this study was to identify the sequences responsible for nuclear targeting. Site-directed mutagenesis and deletion analyses were used to generate a series of golli-green fluorescent protein (GFP) DNA constructs that were transfected into OL and neuronal cell lines to follow localization by confocal microscopy. The results indicated that a 36-residue stretch in the MBP domain is essential for nuclear targeting, and the sequence appears to be a nontraditional localization signal motif. The studies also revealed that overexpression of golli proteins could induce dramatic changes in cell morphology. In OL lines, overexpression of intact golli proteins, or golli peptide alone, caused an increase in the length and number of processes, and the elaboration of membrane sheets. In the neuronal lines, there was a dramatic increase in number and length of extensions. The results, consistent with the timing of golli expression in cells during neural development, suggest that golli proteins may be involved in process formation/extension in OLs and neurons during development. These studies have defined two functional domains in the golli protein. Sequences in the MBP domain target the protein into the nucleus and sequences within the golli domain induce process sheet extension in OLs and neurons. © 2002 Wiley-Liss, Inc.
The myelin basic protein (MBP) gene encodes two families of proteins: the “classic” MBPs and the golli proteins (Campagnoni et al., 1993; Pribyl et al., 1993). In mouse, three golli products have been identified: BG21, J37, and TP8 (Campagnoni et al., 1993; also see Fig. 1A). Unlike the classic MBPs, which are expressed exclusively in myelin-forming cells in the nervous system, the golli proteins are expressed in myelin-forming cells as well as in neurons in the central nervous system (CNS) and peripheral nervous system (PNS; Landry et al., 1996, 1997, 1998; Pribyl et al., 1996) and in macrophages and T-cells in the immune system (Feng et al., 2000). Northern blot analyses, in situ hybridization, and immunohistochemical studies indicate that golli mRNAs and proteins are expressed in the embryonic mouse brain much earlier than the MBP mRNAs and long before myelination. Within the oligodendrocyte (OL), both in vivo and in vitro, golli proteins are detected in the nucleus, cell bodies, and proximal processes, but are not localized within the myelin membrane (Landry et al., 1996; Pribyl et al., 1996; Givogri et al., 2000; Campagnoni and Skoff, 2001). Similarly, golli proteins first appear in many neurons when they are extending processes for migration, establishment of connections and, in the case of OLs, just prior to myelination (Landry et al., 1996, 1997, 1998; Pribyl et al., 1996).

A: Schematic representation of the organization of the mouse mbp gene and the golli and classic MBP (shaded) products derived from the gene. The gene contains two major transcription start sites (tss) that give rise to either the golli products (tss1) or the “classic” MBPs (tss3). Tss2 is a minor start site that gives rise to the M41 MBP transcript, which encodes the 14-kDa MBP. Note that all of the “classic” MBP proteins contain residues 1–36, which are derived from part of exon 5B. B: Diagrammatic scheme of the golli-mbp:green fluorescent protein (GFP) constructs designed to study subcellular targeting of the protein in the glial cell lines. The golli protein was divided into the MBP and golli domain in order to determine whether either regions were responsible for nuclear targeting. Expression of the insert is under the cytomegalovirus (CMV) promoter. C: Mutagenic primers were designed to delete the dibasic lysine-arginine (KR) residues in the MBP domain. Two mutant constructs were generated one in which K5R6 was deleted (MBPΔ1) and the second in which K52R53 was removed (MBPΔ2). D: Two additional MBP:GFP constructs were generated to localize the nuclear targeting sequence within the MBP domain.
Subcellular targeting of the golli protein is rather unusual. In OLs and neurons, golli can be detected in the cell body, nucleus, and processes (Landry et al., 1996; Pribyl et al., 1996). However, within certain subpopulations of neurons, golli is translocated from one cellular compartment to another, which in some cases appears to be developmentally regulated. For example, in the cerebellum, immature granule cells within the external granule cell layer (EGL) target golli into their processes. However, after these cells migrate from the EGL to the internal granule cell layer (IGL), golli is localized solely within the nuclei of the mature granule cells (Landry et al., 1996).
The function of the golli proteins is only beginning to become clarified, and several lines of evidence suggest that it is involved in signaling pathways, and a putative nuclear partner for golli has been identified (Fernandes et al., 2000; Campagnoni and Skoff, 2001). Nuclear cytoplasm shuttling of proteins is a characteristic of many signaling molecules (Bhat, 1995; Hardy and Chaudhri, 1997; Kaffman and O'Shea, 1999; Jordan et al., 2000; Wegner, 2000; Teruel and Meyer, 2000) and is consistent with a signaling role for golli proteins.
Here we describe the identification and characterization of two functional domains of the golli proteins, which may be related to their biological function. We have found that the golli domain can induce dramatic morphological changes in OLs and neurons. These changes include elaboration of membrane sheets in OLs and an increase in number and length of processes in neurons. The MBP domain appears to be essential for targeting the protein into the nucleus. These results are consistent with the notion that the golli proteins may have multiple functions within the cell.
MATERIALS AND METHODS
Construction of the Green Fluorescent Protein (GFP) Clones
DNA insert amplification was performed in accordance to manufacturer's recommendations (GibcoBRL, Rockville, MD). The cycling conditions were as follows: (1) 5 min at 95°C for 1 cycle; (2) 3 min at 95°C, 2 min at 68°C, 2 min at 72°C for 30 cycles. The 5′ and 3′ primers used to generate the BG21, J37, and TP8 reading frames were G5EX25′-CATTAGCTAGCGAATTC ATGGGAAACCACTCTGG-3′, M3END 5′-ATATGAATTCTTGGATCCCGGCTCGGAGCT CACC-3′, J37-GFP 5′-ATATGAATTCTTGGATCCGCGTCTCGCCATGGG- 3′, and TP8-GFP 5′-ATATGAATTCTTGGATCCTCTTGGGTCCGGCCGTGC-3′ (Integrated DNA Technology, Coralville, IA), respectively. The 5′ and 3′ primers used to amplify the Golli specific domain were G5EX25′-CATTAGCTAGCGAATTCATGGGAAACCACTCTGG-3′ and 133END 5′-TGAATTCTTGGATCCCACATCCAGGCCTCCGGAAGC-3′, respectively. To amplify the MBP(1–56), MBP (1–36), and MBP (37–56) sequences, the following primers were used: INT5 5′-CATTAGCTAGCGAATTCATGGCATCACAGAAGAGACC-3′, 5MBP(MLDS) 5′-CATTAGCTAGCGAATTCATGCTTGACTCCATCGGGCGC-3′, 3MEND5B 5′-ATGAATTCTTGGATCCCTTGCCAGAGCCCCGCTTGG-3′, and 3MBP (PRHP) 5′-ATGAATTCTTGGATCCGATGCCCGTGTCTCTGTGCC-3′.
Polymerase chain reaction (PCR) inserts and pEGFP-N3 plasmid (Clontech, Palo Alto, CA) were digested with Bam HI and EcoRI restriction enzymes (New England Biolabs, Beverly, MA) overnight at 37°C. The DNA was pelleted, washed, and resuspended in distilled water. The inserts were then directionally cloned into the GFP vector. The 5′ and 3′ ends of the GFP clones were sequenced with the 5EGFP.SEQ 5′- GGAGGTCTATATAAGCAGAGCTGG-3′ and EGFP-N3.SEQ 5′-CGTCGCCGTCCAGCTCGACCAG-3′ primers, respectively.
Site-Directed Mutagenesis
Site-directed mutations (deletions) were performed on the MBP1–56:GFP clone using the Transformer Site Directed Mutagenesis kit (Clontech) to determine whether either of the two dibasic stretches in the MBP domain are necessary for nuclear targeting. The selection primer used was Not I to EcoRV 5′-GTAAAGCGGATATCCGCGACTCTAGATC ATAATCAGCC-3′. The two mutagenic primers to delete the dibasic residues in the MBP domain were: MBPΔ1 5′-CGAATTCATGGCATCACAGCCCTCACAGCGATCCAAGTACC-3′ and MBPΔ2 5′-GCGGTGACAGGGGTGCGCCCGGCTCTGGCAAGGGATCCATCGC-3′.
Cell Preparation and Transfection
The conditionally immortalized cell lines CN1.4, N19, and N20.1 were grown in Ham's F-12/Dulbecco's modified Eagle's (DME) media (Irvine Scientific, Santa Ana, CA) supplemented with 10% fetal calf serum (FCS; Life Technologies, Gaithersburg, MD) and 200 μg/mL G418 sulfate (Omega Scientific, Tarzana, CA). Cultures were maintained at 34°C with 5% CO2. PC12 cells were grown in Ham's F-12/DME supplemented with 10% new born bovine serum (NBS; Irvine Scientific, CA) and kept at 37°C with 5% CO2.
Cells plated onto poly-D-lysine (PDL)-coated 12-mm glass coverslips were transfected using the Lipofectamine Plus reagent (Life Technologies). Briefly, 900 ng of plasmid DNA was used to transfect 4.5 × 104 cells/coverslip. While the DNA was complexing, the cells were washed three times for 5 min with serum free media. The complexed DNA mixture was then applied to the coverslips and incubated at 34°C for 6 hours. The samples were washed 3 times with media supplemented with 10% FCS and subsequently incubated at 34°C for 3 days prior to fixation.
Cells were briefly rinsed three times with 1× phosphate-buffered saline (PBS) and then fixed in freshly prepared 2% paraformaldehyde in PBS for 40 min at room temperature. Following fixation, the cells were washed three times for 5 min in PBS and stored at 4°C.
Immunofluorescence Cytochemistry
The protocol for the immunofluorescence studies was in accordance to that mentioned by Bongarzone et al. (1996) with the following modifications. Fixed cells were permeabilized with 0.1% Triton X-100 in PBS for 10 min at room temperature followed by three 5-min washes in PBS. Samples were incubated in a blocking solution (5% goat serum in PBS) for 1 hour at room temperature with mild agitation. The coverslips were then probed with anti-Golli, galactocerebroside (GC), proteolipid protein (PLP), and MBP antibodies (Golli, 1:1,000; 01, 1:20; AA3, no dilution; MBP, 1:700 in 0.1% blocking solution in PBS) overnight at 4°C with mild shaking. The following day, the coverslips were allowed to reach room temperature prior to the following procedures. The cells were washed three times for 5 min in PBS then probed for 2 hours at room temperature with the secondary antibody conjugated to Texas Red (1:800; Jackson Immunoresearch Laboratories, West Grove, PA). The coverslips were washed three times for 10 min in PBS, once in distilled water, and then mounted onto glass slides with Aquamount (Lerner Laboratories, Pittsburgh, PA).
Fluorescent images were obtained using a Leica upright DMRXA and a Leica inverted DMIRB microscope. Morphometric analysis was performed using Image Pro Plus software (Media Cybernetics; Silver Spring, MD).
RESULTS
Subcellular Localization and Translocation of the Golli Protein In Vitro
Immunocytochemical analyses of the two conditionally immortalized mouse OL cell lines, N19 and N20.1 (Foster et al., 1993; Verity et al., 1993) shows that golli is localized primarily in the nucleus (Fig. 2A,E) and to a lesser extent, in the cytoplasm and processes. We felt these two lines would be good models for studying nuclear transport of golli since its nuclear localization implies the lines must have the machinery to transport golli to the nucleus, and because these cells would be more relevant to neural tissue than non-neural transformed cells.
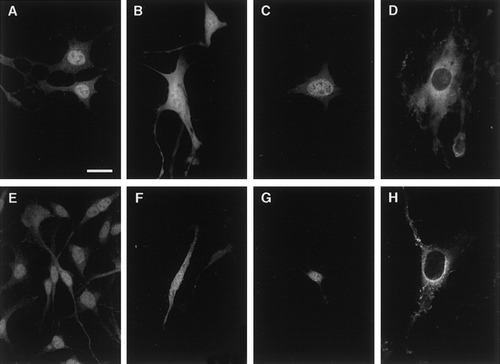
Subcellular localization of golli-mbp proteins in oligodendroglial cell lines N19 (A,B,C,D) and N20.1 (E,F,G,H). Nontransfected cells (A,E) show strong nuclear staining of golli immunoreactivity. Confocal images of cells transfection with the nonmodified GFP vector showed an even distribution of the protein in the processes, cell body, and nucleus (B,F), whereas nuclear targeting of the chimeric protein was observed in cells transfected with the MBP1–56:GFP construct (C,G). Cells transfected with the Golli:GFP construct localized the chimeric protein into the cytoplasm and in particular within the perinuclear region (D,H). A change in cell morphology was observed in both the transfected N19 and N20.1 cells that was not seen in either the nonmodified GFP vector or the MBP1–56:GFP transfectants. Scale bar = 10 μm.
To examine the subcellular localization of the golli protein, a series of constructs were prepared with portions of the golli proteins fused to the green fluorescent protein (GFP), to permit the easy localization of the transfected protein in the cell line (Fig. 1B). The DNA constructs were transiently transfected into the N19 and N20.1 cell lines, and targeting of the chimeric proteins was visualized using confocal microscopy.
The MBP Domain Within the Golli Protein is Responsible for the Targeting of the Golli Proteins Into the Nucleus of OL Cell Lines
Figure 2 illustrates confocal optical sections of N19 and N20.1 transfected with the golli:GFP and MBP:GFP constructs maintained at 34°C. Figure 2B and F shows that in N19 and N20.1 cells transfected with the vector alone, the GFP protein was evenly distributed throughout the cell. This is consistent with the findings that small molecular weight proteins, such as the 27-kDa GFP molecule, passively diffuse through cellular compartments (Gorlich and Mattaj, 1996; Gorlich and Kutay, 1999; Pedraza and Colman, 2000). When both lines were transfected with the MBP1–56:GFP construct the chimeric protein was targeted primarily into the nucleus (Fig. 2C,G). In contrast, in cells transfected with the golli:GFP construct, the chimeric protein was localized in the cytoplasm, particularly within the perinuclear region (Fig. 2D,H).
To investigate further which residues are necessary for nuclear targeting of the golli proteins, we identified basic amino acid sequences that could serve as potential nuclear localization signals (NLS). Computer analyses of MBP residues 1–56, the MBP domain in BG21, failed to identify any putative bipartite motifs, a common NLS found in more than 50% of known nuclear proteins (Dingwall and Lasky, 1991; Makkerh et al., 1996). However, two dibasic single clusterlike stretches were identified. To determine whether these sequences were necessary for nuclear targeting, site-directed mutagenesis of the MBP1–56:GFP clone was used to generate two constructs in which either K5R6 or K52R53 was deleted. These were designated MBPΔ1 and MBPΔ2, respectively (Fig. 1C). These mutant constructs were transfected into the N20.1 line and examined by confocal microscopy.
Figure 3C shows that deletion of the K5R6 residues (MBPΔ1) did not abolish nuclear targeting in the N20.1 line. In the transfected cells, the chimeric protein was localized in the nucleus. Similar results were also observed in cells transfected with the MBPΔ2 and the control construct, MBP1–56:GFP (Fig. 3D,B). Transfection of the N19 cell line with these constructs produced identical results (data not shown).
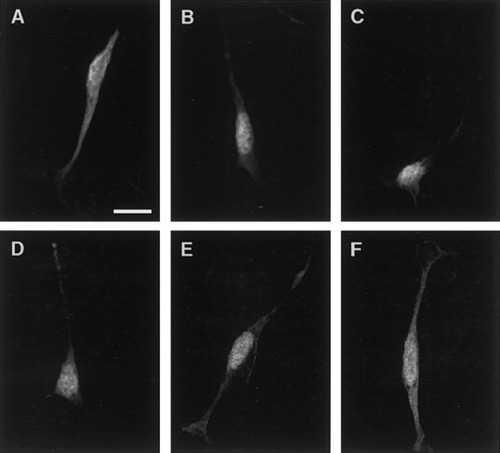
Confocal images of N20.1 cells transfected with the site directed mutant constructs show that deletion of the dibasic residues did not abolish nuclear targeting. N20.1 transfected with the nonmodified GFP vector shows an even distribution of GFP protein into the processes, cell body, and nucleus (A). The MBP (1–56) protein was targeted to the nuclei of transfected cells (B). Deletion of MBP residues K5R6 did not abolish nuclear accumulation (C) nor was nuclear targeting disrupted by the deletion of residues K52R53 (D). Nuclear targeting sequence in the MBP domain is within the first 36 amino acids. Nuclear accumulation of the chimeric protein was observed in cells transfected with both the MBP1–36:GFP (E). In contrast, cells transfected with the MBP 37–56:GFP construct did not target the protein solely within their nuclei but rather evenly distributed the protein into the processes, cell body, and nucleus (F). Scale bar = 10 μm.
To refine further the region within the MBP domain that contains the sequences necessary for nuclear targeting, two sets of PCR primers were designed and used to generate constructs containing MBP residues 1 through 36, and 37 through 56, respectively (Fig. 1D), fused to GFP. In the N20.1 cells transfected with the MBP1–36:GFP construct the protein was localized in the nucleus (Fig. 3E). In contrast, in the cells transfected with the MBP 37–56:GFP construct the chimeric protein (Fig. 3F) was distributed throughout the cell, similar to what was seen in cells transfected with the GFP vector alone (Fig. 3A).
Cells That Overexpress Golli Proteins Undergo Major Morphological Changes
During the course of identifying sequences in the golli protein necessary for nuclear targeting, we observed that the N20.1 line underwent significant morphological changes after transfection with the golli:GFP construct. These morphological changes were not seen in cells transfected with either the control GFP vector or the MBP:GFP constructs, suggesting that the golli domain was responsible for this. To examine this further, the full-length BG21, J37, and TP8 reading frames were cloned into the GFP vector and then transfected into the N19 and N20.1 lines.
Figure 4 illustrates fluorescent images of N19 and N20.1 transfected with the control GFP vector, and the BG21:GFP, J37:GFP, and TP8:GFP constructs at 34°C. No morphological changes were induced in either the N19 cell line (Fig. 4A) or the N20.1 cell line (Fig. 4E) transfected with the control GFP vector. The cells retained the appearance of nontransfected cells. After these cells were transfected with the BG21:GFP construct, the morphology of both OL lines became flatter with thickened processes and they elaborated large membranous sheets (Fig. 4B,F). Similar morphological changes were observed in cells transfected with the J37:GFP and TP8:GFP constructs (Fig. 4C,G,D,H), indicating that the morphological changes and membrane sheet elaboration could be induced by overexpression of all the golli isoforms of the MBP gene.
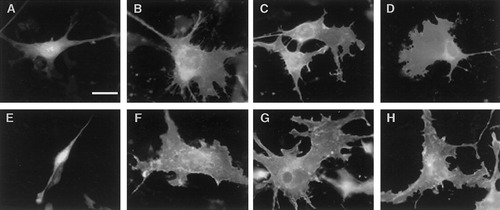
Elaboration of membrane sheets in the N19 and N20.1 cell lines transfected with the full-length golli products. Fluorescent images of N19 cell transfected with the nonmodified GFP vector did not undergo any dramatic changes in cell morphology (A), whereas cells transfected with the full length BG21 (B), J37 (C) or TP8 (D) appeared broader and had elaborated extensive membrane sheets. N20.1 cells transfected with the GFP vector did not show any significant membrane sheet formation (E). However, cells overexpressing BG21 elaborated broad membrane sheets and these cells appeared flatter than the GFP control (F). Similar results were seen in cells transfected with J37:GFP (G) and TP8:GFP (H). Scale bar = 10 μm in A,E; 25 μm in B,C,D,F,G,H.
These morphological changes were confirmed by quantitative measurements using an imaging analysis program. Figure 5 compares the mean surface area of N20.1 cells transfected with the various golli constructs. The surface area of the N20.1 cells transfected with BG21, J37, and TP8 was significantly larger (*P <.00001) than cells transfected with the MBP portion of BG21 or with the control GFP vector.
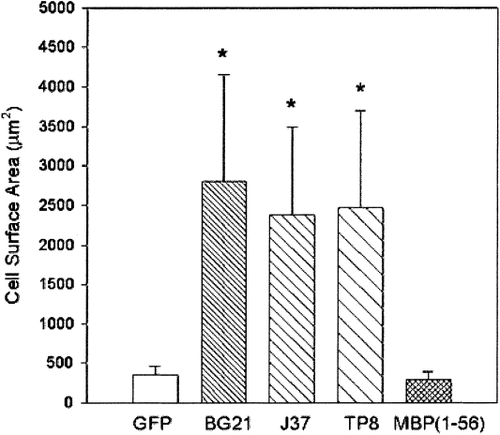
Morphometric analyses of the mean ± S.E.M. surface area of N20.1 cells transfected with BG21, J37, TP8, or MBP(1–56) illustrates that N20.1 cells overexpressing the golli-specific sequence, or full-length BG21, J37, or TP8, showed a significant increase in surface area not seen in cells transfected with the nonmodified GFP vector or the MBP(1–56):GFP construct (n = 50; analysis of variance [ANOVA], *P < 0.00001).
To determine whether the effects of overexpression of golli proteins on cell morphology were unique to OLs we performed similar experiments on two “neuronal” cell lines. One of these was a conditionally immortalized cortical neuronal cell line (CN1.4) produced in this laboratory (Bongarzone et al., 1998) and the rat pheochromocytoma PC12m line. Both lines were transiently transfected with the BG21:GFP construct and examined for morphological changes. Shown in Figure 6 is a series of fluorescent micrographs illustrating the types of morphological changes induced as a consequence of overexpression of BG21. In both lines there was an increase in the number and length of processes in the transfected cells. The processes in the PC12m cells were thicker, longer, and branched (Fig. 6B,C). In contrast, cells transfected with the nonmodified GFP vector exhibited few short thin processes (Fig. 6A). Extension of processes in PC12 cells is reminiscent of the type of morphological changes induced in the presence of nerve growth factor (NGF). The CN1.4 cells elaborated long thick processes (Fig. 6E,F) not seen in the GFP transfected cells (Fig. 6D).
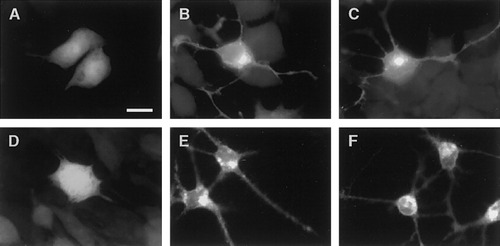
Fluorescent images show that overexpression of BG21 results in the extension of processes in the neuronal cell lines PC12m and CN1.4. Control experiment in which PC12m cells transfected with the nonmodified GFP vector appear not to have undergone any significant change in cell morphology (A). In contrast, cells transfected with the BG21:GFP construct showed an increase in number and length of extensions (B,C). Similarly, CN1.4 cells transfected with the GFP vector did not show any change in morphology (D), whereas the processes in CN1.4 cells transfected with the BG21:GFP construct appear thicker and longer (E,F). Note that the PC12m cells were not grown in the presence of nerve growth factor (NGF). Scale bar = 10 μm.
The Membrane Sheets Elaborated by Transfected OL Cell Lines are not “Myelin-Like”
Because the morphological changes induced in the OL cell lines were similar to OLs grown in primary culture (Fig. 7A,B,D,E,G,H) we wanted to determine whether the membranous sheets elaborated by the transfected cells had characteristics of myelin. If this were so, then the golli proteins might be acting to induce the lines to differentiate. Immunocytochemical analyses were performed with standard markers of myelin to determine whether overexpression of the golli proteins in the N19 and N20.1 lines resulted in a differentiated phenotype. The membranes were not positive for any of the myelin markers examined. Examples of some of these are shown for cells stained for 01 in transfected N19 cells (Fig. 7C), and for MBP and PLP in transfected N20.1 (Fig. 7F,I).

Immunocytochemical analyses show that the elaborated sheets are not immunopositive for galactocerebroside (GC), MBP, or proteolipid protein (PLP) and are not myelin-like. OLs, 21 Days in vitro (DIV), showed intense GC staining (A). In contrast, the membrane sheets elaborated in N19 cells transfected with the BG21:GFP construct (B) remained negative for GC (C). Oligodendrocytes (OLs) at 21 DIV were also immunopositive for MBP (D), whereas N20.1 cells, expressing the BG21:GFP construct (E), did not stain for MBP (F). PLP immunoreactivity was clearly present in OLs at 21 DIV (G). However, N20.1 cells transfected with the BG21:GFP construct (H) did not show any PLP immunoreactivity (I). Scale bar = 15 μm in A; 16.7 μm in D; 10 μm in G; 30 μm in B,C,E,F,H,I.
DISCUSSION
In this report, we have shown that golli proteins contain two functional domains: an MBP domain, responsible for targeting the proteins into the nucleus; and a golli domain, which can induce dramatic morphological changes in both OLs and neurons.
Our data indicate that the nuclear localization signal for the golli proteins is found within the first 36 amino acids of the MBP domain, corresponding to exon 5b (or exon 1 of the MBP gene using the old numbering system). We did not identify a conventional NLS within the MBP domain of the golli proteins, so the mechanism for nuclear targeting of the golli proteins is probably regulated by sequences that do not involve traditional NLS motifs (Dytrych et al., 1998; Sherman and Brophy, 2000).
The MBP residues 1 through 36 contain numerous potential PKC phosphorylation sites. This is interesting because MBP proteins have been reported to be phosphorylated, at least partially, in vivo (Ulmer and Braun, 1986a,b) and in vitro (Miyamoto et al., 1974; Miyamoto and Kakiuchi, 1974). Pedraza et al. (1997) has shown that the addition of 12-O-tetradecanoyl phorbol 13-acetate (TPA), a potent activator of most PKC isoforms, can abolish nuclear targeting of the 21.5-kDa MBP isoform transfected into HeLa cells. Therefore it is possible that the phosphorylated state of the MBP domain in the golli products contributes to determining the subcellular location of the protein.
The minor isoforms of the classic MBPs, 21.5 kDa and 17 kDa MBP, have been observed in nuclei of OLs in the developing mouse brain (Allinquant et al., 1991; Hardy et al., 1996; Staugaitis et al., 1996). It has been shown that MBP sequences encoded by exon 6 (old exon 2) are necessary for this nuclear translocation in the classic MBP isoforms. However, exon 6 is not present in the golli products, so the golli proteins utilize a different NLS than the minor MBP isoforms. Since the golli NLS is present in all the classic MBP isoforms, then why are they not targeted to the nucleus? The context in which the NLS is found and other factors influence whether or not it functions as a NLS (Tagawa et al., 1995; Yoneda, 2000), and the structures of the golli and classic MBPs are very different. Perhaps, more importantly, targeting of the classic MBPs to the myelin membrane is achieved by polyribosome translocation, which effectively bypasses the effects of nuclear targeting signals in these molecules (for recent review see Landry and Campagnoni, 1998).
The golli domain alone was responsible for the ability of the golli proteins to induce the elaboration of processes and membranous sheets when overexpressed in OL and neuronal cell lines. Unlike the J37 and BG21 golli isoforms, which contain the entire 133 aa golli domain, TP8 contains only the first 47 aa of this sequence and is lacking any MBP domain. Thus, these data suggest that the first 47 residues of the golli domain are sufficient to induce the observed changes in cell morphology.
The findings that overexpression of the golli proteins induced elaboration of membrane sheets in the OL cell lines is significant. While the mechanism for membrane sheet elaboration in OLs is not fully understood, in vitro studies indicate that sheet formation involves the activation of PKA and PKC (Vartanian et al., 1986; Althaus et al., 1990, 1991; He and McCarthy, 1994; Yong et al., 1994; Bhat, 1995; Yong and Oh, 1997). Furthermore, the extracellular signal-regulated protein kinases (ERKs), members of the mitogen-activated protein kinase (MAPK) family, and Fyn, a member of the Src tyrosine kinase family, have been shown to have an important role in the initiation of process formation in OLs (Umemori et al., 1994; Bhat, 1995; Stariha et al., 1997; Osterhout et al., 1999; Seiwa et al., 2000). Therefore, membrane sheet formation in the N19 and N20.1 lines transfected with the golli constructs suggests that the golli proteins might be involved in some way with a signal transduction pathway involved in this process.
Similarly, overexpression of golli proteins in PC12m cells led to the induction of processes, similar to the induction seen with NGF. Like the OL lines, golli is also expressed abundantly in PC12m cells. In contrast to sheet extension in OLs, neurite extension in PC12 cells has been well characterized in vitro (Tischler and Greene, 1975, 1978, 1980). Numerous studies have shown that cells treated with NGF differentiate and extend neurites (Greene and Tischler, 1976; Szeberenyi and Erhardt, 1994; Friedman and Greene, 1999), mediated by the binding of NGF to the TrkA receptor, tyrosine kinase receptor (Barbacid, 1994; Saltiel and Decker, 1994; Greene and Kaplan, 1995; Kaplan, 1998; Friedman and Greene, 1999). Activation of the receptor leads to the initiation of several signal transduction pathways, including the Ras-MAPK pathway which is necessary for neurite extension (Pang et al., 1995; Robinson and Cobb, 1997; Korhonen et al., 1999). Interestingly, Katoh et al. (2000) have shown that overexpression of RhoG in PC12 can also induce neurite extension in the absence of NGF. RhoG is a member of the Rho GTPase family that can serve as activators of several signal transduction pathways (Hall, 1998; Bar-Sagi and Hall, 2000). These data suggest that overexpression of a modulator within the signaling pathway can bypass NGF activation to initiate neurite extension. Overexpression of golli might be acting in a similar fashion in PC12m cells to induce process extension.
Our results suggest that golli proteins may be involved in process elaboration/extension in both OLs and in neurons. In OLs, golli is expressed very early, prior to myelin formation, when the cells are beginning to extend processes (Pribyl et al., 1996). These in vitro results suggest that myelination may be a multiple stage process: extension of cell processes, the beginning of sheet elaboration, followed rapidly by delivery of myelin components to the membrane. Since golli is not a component of myelin, it may be involved in setting up process/sheet formation prior to the delivery of the myelin components to the growing membrane.
In this study, we report our findings that the golli protein contains two functional domains. Sequences located in the MBP domain targets the protein into the nucleus while the golli domain induces cell-specific morphological changes. Although the function of the golli proteins is not yet known, the present studies suggest that golli may be involved in signaling mechanisms within the cell and that this activity resides within the golli domain of the molecule.




