Sensitive and Visualized Detection of Hantavirus Using CRISPR/Cas12a Based on AutoCORDSv2 Design
Changsheng He, Wei Zhu, Xin Zhang, and Wei Wu contributed equally to this study.
ABSTRACT
In recent years, detection technologies based on the CRISPR/Cas12a method have been extensively utilized in the fields of nucleic acid, enzyme, and macromolecule detection, thereby reinforcing their significant role in the detection landscape. Enhancing the simplicity of design, efficiency, and automation of the CRISPR/Cas12a detection system is essential for advancing its application in diagnostics. Recently, we developed an automated CRISPR/Cas12a design system named AutoCORDSv2. This system can process published genomic sequences of pathogenic bacteria in a high-throughput manner and automatically generate conserved and highly specific crRNA sequences, along with primer sequences for target amplification. This capability facilitates the specific and precise design of the CRISPR/Cas12a detection system. In this study, crRNAs targeting the Hantaan virus (HTNV) and Seoul virus (SEOV), as well as RT-PCR primers and RT-RPA primers, were designed using AutoCORDSv2. The experimental results demonstrated that the CRISPR/Cas12a system, automatically designed by AutoCORDSv2, was specific for the detection of both the HTNV and SEOV, with no cross-reactivity observed with other pathogens. The detection sensitivity reached 6 copies/μL (equivalent to 111 copies per amplification reaction), whether measured by a microplate reader or directly observed with the naked eye. The detection results for 50 samples were consistent with those obtained from commercial RT-qPCR kits, indicating high precision. Furthermore, the CRISPR/Cas12a system designed by AutoCORDSv2 can also be utilized for the development of a single-tube detection system with a sensitivity of 42 copies per reaction. This system combined with a 5-min extraction step and RT-RPA, further underscoring its potential for application.
1 Introduction
Hantaviruses are classified as negative-strand RNA virus characterized by their enveloped structure and segmented genome, which comprises L, M, and S segments. These segments encode the L polymerase protein, glycoproteins G1 and G2, and the nucleocapsid protein, respectively. Hantaviruses exhibits a nearly global distribution and has the potential to cause significant morbidity and mortality in humans [1]. Hantaviruses are responsible for diseases such as hantavirus cardiopulmonary syndrome (HCPS), which is endemic in the Americas, and hemorrhagic fever with renal syndrome (HFRS), which is endemic in Europe and Asia; however, these viruses do not induce any syndromes in their primary host, the mouse. HTNV and SEOV are the two most common hantaviruses that cause HFRS, especially in China [2].
The predominant technologies utilized for the detection of HTNV and SEOV encompass sequencing, enzyme-linked immunosorbent assay (ELISA), and reverse transcription quantitative polymerase chain reaction (RT-qPCR). Recently, to mitigate reliance on large and costly instruments, several methods for isothermal nucleic acid detection have been developed. These methods include isothermal amplification technologies, such as reverse transcription loop-mediated isothermal amplification (RT-LAMP) [3-5]. Notably, CRISPR-based nucleic acid detection technology warrants attention, as it advances diagnostics toward point-of-care applications [6]. In CRISPR/Cas12a (also called Cpf1) or CRISPR/Cas13a (also called C2c2)-based detection, the Cas12a or Cas13a proteins cleave the target DNA or RNA that is complementary to the CRISPR RNA (crRNA) [7-9]. Subsequently, their trans-cleavage activity for nonspecific single-stranded DNA (ssDNA) or single-stranded RNA (ssRNA) is activated [10-14]. Ultimately, the ssDNA or ssRNA reporter is cleaved, resulting in the release of detectable signals. The entire process of CRISPR detection can be conducted at 37°C, enhancing its utility for point-of-detection applications [15-24].
Numerous CRISPR-based methodologies have been developed; however, an integrated design tool for crRNA and primers for the amplification of crRNA target sites remains unavailable. In our recent study, we introduced AutoCORDSv2, a tool for the automated design of CRISPR/Cas12a crRNAs and primers [25]. In this study, we employed AutoCORDSv2 to design crRNAs and primers targeting the S gene of HTNV and SEOV. Our evaluation demonstrated that the crRNAs and primers designed by AutoCORDSv2 exhibited both efficiency and specificity for the detection of HTNV and SEOV, achieving a sensitivity of 6 copies/μL without cross-reactivity with other viruses. To further streamline and expedite the detection process, we also developed a one-pot Cas12a detection system for HTNV and SEOV in this study. Additionally, AutoCORDSv2 proves to be a valuable and user-friendly tool for the automated design of conserved and efficient crRNAs and primers, which can be utilized in the detection of HTNV, SEOV, or other pathogens.
2 Methods
2.1 Materials and Instruments
One-Step reverse-transcription PCR (RT-PCR) Kit and Green Taq Mix were purchased from Vazyme Biotech. RT-RPA Kit was purchased from Amplification Future. LbCas12a, 10 × NEBuffer r2.1, and 10 × NEBuffer r3.1 were purchased from NEB. GeneJET Plasmid Miniprep Kit, T7 TranscriptAid T7 Transcrip Kit and RiboLock RNase Inhibitor were purchased from ThermoFisher. RNA Clean & Concentrator-100 was purchased from ZYMO RESEARCH. Gel Extraction Kit was purchased from Omega Biotech. Oligonucleotides and DNA were synthesized by Sangon Biotech. Nuclease-free PCR tubes and Pipet tips were purchased from Axygen. 384-well microplates were purchased from Greiner. Nucleic acids were quantified using Quawell Q6000 Micro Volume Spectrophotometer. PCR and thermostatic reaction were conducted with BIO-RAD T100 Thermal Cycler. Fluorescence signals were recorded with Agilent BioTek Synergy H1 Multi-Mode Microplate Reader and visualized with TOUHE blue light glue cutter THBC-470 or BLT GelView 5000Plus.
2.2 crRNA Design and Transcription
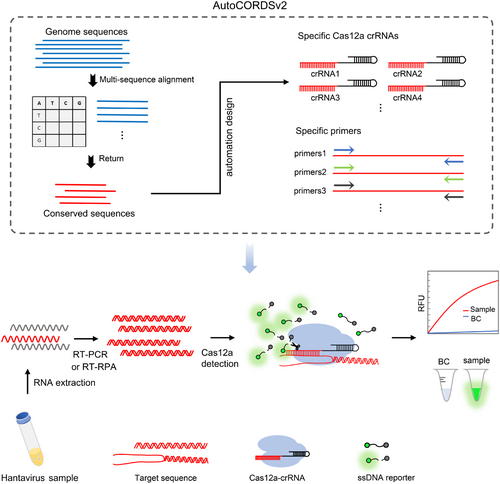
The crRNAs used in this study were automatically designed by AutoCORDSv2 (Figure 1), which was developed for rapid selection of crRNA and primer in previous study [25]. After inputting the genomic sequences of the virus, AutoCORDSv2 automatically activates the conservative analysis module to perform a conservative alignment using either the default threshold (e.g., 99%) or a customized conservative threshold. This process allows for the rapid identification and filtering of specific crRNA and primers within the highly conserved regions. In this study, a total of 189 HTNV S gene sequences and 85 SEOV S gene sequences were retrieved from the Bacterial and Viral Bioinformatics Resource Center (BV-BRC, https://www.bv-brc.org/). These sequences were subsequently analyzed using AutoCORDSv2 to identify HTNV and SEOV-specific as well as conserved crRNAs.
After the design of crRNAs, as in previous studies [25-27], 60 nt oligonucleotides containing a T7 promoter, crRNA scaffold, and crRNA were synthesized (Table 1 and Supporting Information S1: Table S1). During the crRNA transcription process, oligo-F and oligo-R were annealed using NEBuffer r3.1 to form the DNA template for transcription. Subsequently, crRNAs were obtained using the T7 TranscriptAid T7 Transcrip Kit according to the standard protocol. The transcription products were then treated with DNase I and purified using the RNA Clean & Concentrator-100 kit. Finally, the purified crRNAs were quantified and stored at −80°C before use.
| crRNA with PAM (5′ > 3′) | Conserve score | CG |
|---|---|---|
Han-crRNA1 TTTG CACTATTATTATCAGGGGAA |
0.965828924 | 35% |
Han-crRNA2 ATTC CCACCCATAAATGTTCCCAT |
0.970458554 | 45% |
Han-crRNA3 TTTA TGGGTGGGAATCATTACTCA |
0.967813051 | 45% |
Seo-crRNA1 TTTG ATTGTATTGAAGCTGCGACA |
0.988725490 | 35% |
Seo-crRNA2 TTTT GATTGTATTGAAGCTGCGAC |
0.987745098 | 45% |
Seo-crRNA3 CTTC TGGCGTGCTATCACAAGCTG |
0.987254901 | 55% |
Seo-crRNA4 CTTA AACAAGAGGGCACTGCATGA |
0.986764705 | 49% |
2.3 Target DNA and RNA Preparation
In the preparation of target DNA, conserved sequences containing specific crRNA targets for HTNV and SEOV were synthesized and cloned into the pUC57 vector, and were named as pUC57-Han and pUC57-Seo (Supporting Information S1: Table S2). For target RNA preparation, primers containing a T7 promoter were utilized to amplify the conserved sequences in pUC57-Han and pUC57-Seo using Green Taq Mix (Supporting Information S1: Table S2). Subsequently, target RNAs were obtained, purified, and quantified as described above, using the PCR products as templates for transcription.
2.4 crRNA Screen Assay
Three crRNAs targeting HTNV and four crRNAs targeting SEOV were screened to identify the optimal candidates. For the HTNV crRNA screening assay, the following components were mixed to achieve a total reaction volume of 20 μL: 2 μL of 10× NEBuffer r2.1, 0.5 μL of RiboLock RNase Inhibitor, 50 nM of LbCas12a, 100 nM of crRNA (Han-crRNA1, Han-crRNA2, or Han-crRNA3), 500 nM of ssDNA reporter (FAM-TTATT-BHQ1), 1 ng of pUC57-Han target DNA, and nuclease-free water. For the SEOV crRNA screening assay, the same procedure was followed, using 2 μL of 10× NEBuffer r2.1, 0.5 μL of RiboLock RNase Inhibitor, 50 nM of LbCas12a, 100 nM of crRNA (Seo-crRNA1, Seo-crRNA2, Seo-crRNA3, or Seo-crRNA4), 500 nM of ssDNA reporter, 1 ng of pUC57-Seo target DNA, and nuclease-free water to achieve a total volume of 20 μL. The mixtures were then incubated at 37°C, and the Relative Fluorescence Units (RFU) were recorded every 3 min for a duration of 1 h.
2.5 Sensitivity Assay
In the sensitivity assay, the target RNA for HTNV and SEOV was diluted to concentrations of 6000, 600, 60, 6, 1, 0.6, and 0 copies/μL (Blank Control). RT-PCR reactions were subsequently performed to prepare the activators for the CRISPR/Cas12a reaction using a One-Step RT-PCR Kit.
For HTNV, the preamplification involved a 50 μL RT-PCR reaction containing the following components: 25 μL of 2× One Step Mix, 2.5 μL of One Step Enzyme Mix, 0.4 μM each of Han-RT-PCR-primer-F (5′-GCACAGAGCTTGATTGATGTCA-3′) and Han-RT-PCR-primer-R (5′-TAAACCCACCCACAACGGAT-3′), and 18.5 μL of a serial concentration gradient of HTNV RNA. The amplification program was as follows: 25 min at 50°C for reverse transcription; 2 min at 94°C for initial denaturation; 30 cycles of 10 s at 94°C for denaturation, 30 s at 55°C for annealing, and 10 s at 72°C for extension; followed by a final extension of 3 min at 72°C. The Cas12 detection reactions were then performed as follows: a 20 μL reaction volume containing 8 μL of RT-PCR products, 2 μL of 10× NEBuffer r2.1, 0.5 μL of RiboLock RNase Inhibitor, 50 nM of LbCas12a, 100 nM of Han-crRNA1, 500 nM of ssDNA reporter, and nuclease-free water. The mixtures were incubated at 37°C, and the RFU were recorded every 5 min for a duration of 1 h. Visualization was achieved using a TOUHE blue light gel cutter THBC-470 or a BLT GelView 5000Plus after 1 h.
For SEOV, during the preamplification step, a 50 μL RT-PCR reaction was prepared containing the following components: 25 μL of 2× One Step Mix, 2.5 μL of One Step Enzyme Mix, 0.4 μM each of Seo-RT-PCR-primer-F (5′-GGAGGAAATCCAGAGAGAAAT-3′) and Seo-RT-PCR-primer-R (5′-CCAGTGTATTCCCGTAGCTTA-3′), and 18.5 μL of a serial concentration gradient of SEOV RNA. The amplification program was conducted as previously described. Subsequently, a 20 μL Cas12a reaction was prepared, which included 8 μL of RT-PCR products, 2 μL of 10× NEBuffer r2.1, 0.5 μL of RiboLock RNase Inhibitor, 50 nM of LbCas12a, 100 nM of Seo-crRNA3, 500 nM of ssDNA reporter, and nuclease-free water. The mixtures were then incubated at 37°C, and the RFU were recorded every 5 min for a duration of 1 h. Visualization was performed using a TOUHE blue light gel cutter THBC-470 or a BLT GelView 5000Plus after 1 h.
2.6 Specificity Assay and Sample Detection
Nucleic acid samples from two dengue virus (DENV) strains, two Zika virus strains, and one severe fever with thrombocytopenia syndrome virus (SFTSV) were utilized to conduct a specificity assay. Additionally, RNA samples from 50 lung specimens of wild rats were employed for sample detection. These samples were collected from Center for Disease Control and Prevention of Guangdong Province and processed according to strict standard procedures. All of these samples were used for Cas12a-based detection of HTNV and SEOV. The preamplification and Cas12a reaction were performed as described in Section 2.5. Visualization of the detection results was conducted using a TOUHE blue light gel cutter THBC-470 after a 1 h Cas12a reaction.
2.7 One-Pot RT-RPA-Cas12a for HTNV and SEOV Detection
RPA primers were designed using AutoCORDSv2 (Supporting Information S1: Table S1), and the optimal primer was screened for subsequent studies using agarose gel electrophoresis. The one-pot RT-RPA-Cas12a reaction was performed in a single Axygen tube. In brief, 27 μL of the preamplification RT-RPA reaction was added to the bottom of the tube, which contained the following components: 14.7 μL of A buffer with enzyme particles, 0.37 μM each of Han-RPA-primer-1F and Han-RPA-primer-1R or Seo-RPA-primer-1F and Seo-RPA-primer-1R, 7.05 μL of sample RNA, 500 nM of ssDNA reporter, and 1.25 μL of B buffer. To avoid cross-contamination, 25 μL of mineral oil was added to the top of the RT-RPA system. The 19 μL of Cas12a reaction mixture was added to the lid of the tube, containing 2 μL of 10× NEBuffer r2.1, 0.5 μL of RiboLock RNase Inhibitor, 1 μL of LbCas12a (1 μM), 2 μL of Han-crRNA1 (1 μM) or Seo-crRNA3 (1 μM), and nuclease-free water. After incubating for 30 min at 37°C to allow for sufficient amplicon production, the Cas12a reaction mixture was mixed with the RT-RPA mixture by a brief spin, and a 30-min reaction was performed at 37°C for Cas12a detection. Finally, the visualization of the detection results was conducted using a TOUHE blue light gel cutter THBC-470.
To assess the sensitivity of one-pot RT-RPA-Cas12a method for HTNV and SEOV, the target RNA for HTNV and SEOV was diluted to concentrations of 6000, 600, 60, 6, 0.6, and 0 copies/μL (Blank Control). One-pot RT-RPA-Cas12a reactions were performed as described above. For the specificity assay and sample detection, nucleic acid samples outlined in Section 2.6 were analyzed using one-pot RT-RPA-Cas12a reaction as detailed above.
For RNA extraction-free, samples were lysed with HCl solution, which has been reported used for SARS-CoV-2 lysis [28]. Specifically, four SEOV and one HTNV sample homogenates (National Institute for Viral Disease Control and Prevention, China CDC) were mixed with 1 M HCl at a 1:9 volume ratio (HCl: Sample) and allowed it to react for 5 min. The reaction was then terminated by adding 1 M NaOH in the same ratio, and the lysate was used directly for single-tube RT-RPA-Cas12a detection.
2.8 Statistical Analysis
Statistical analyses in this study were conducted using GraphPad Prism. All replicate experiments consisted of three repetitions. Uncertainties in mean values are reported as standard errors. Statistical significance was evaluated using one-way analysis of variance and multiple comparisons. Significance levels were defined as follows: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, and ns indicates no significance.
3 Result
3.1 Efficient and Conserved crRNAs for Cas12a-Based Detection of HTNV and SEOV Were Automatically Designed Using AutoCORDSv2
In a previous study [25], we developed an automated designer called AutoCORDSv2 (https://github.com/duao42/AutoCORDSv2) for CRISPR/Cas12a crRNA rapid screening. In this study, we set out to create a CRISPR/Cas12a method for the detection of HTNV and SEOV based on crRNAs and primers designed automatically by AutoCORDSv2 (Figure 1). The S gene sequences of HTNV and SEOV were input into AutoCORDSv2, which returned the corresponding crRNAs. Among these crRNAs, three with a conserve score higher than 0.965 were selected for HTNV targeting, while four with a conserve score higher than 0.986 were chosen for SEOV targeting (Table 1).
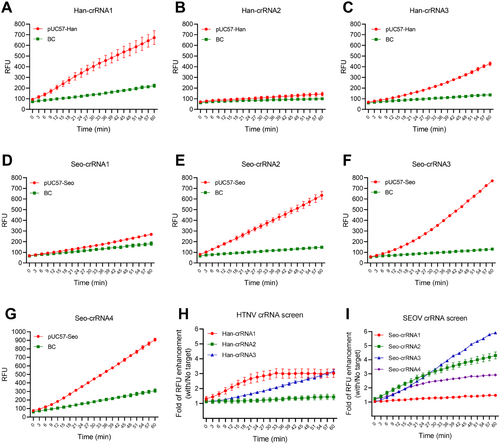
We next conducted an in vitro trans-cleavage assay to identify the optimal crRNAs for the detection of HTNV and SEOV. In the screening of crRNAs for HTNV, results indicated that Han-crRNA1 was the most effective crRNA for activating Cas12a and cleaving the ssDNA reporter in the presence of target DNA (Figure 2A–C, and Supporting Information S1: Figure S1A). For SEOV, the results showed that ssDNA reporters were significantly cleaved when Seo-crRNA2, Seo-crRNA3, and Seo-crRNA4 were complexed with Cas12a and target DNA, in contrast to the minimal increase in fluorescence observed with Seo-crRNA1 (Figure 2D–G). When the RFU was background corrected, selecting the optimal crRNA for SEOV proved to be challenging (Supporting Information S1: Figure S1B). Therefore, the folds of RFU enhancement with and without the target were calculated. The results indicated that Seo-crRNA3 was the most effective, as it exhibited the maximum RFU enhancement after a 30 min reaction (Figure 2I). Additionally, Han-crRNA1 demonstrated the highest RFU enhancement among the three crRNAs within a 60 min reaction (Figure 2H). These results indicate that the conserved crRNAs auto-designed by AutoCORDSv2 exhibit a high efficiency in activating Cas12a, even when only 1 ng of target DNA is present.
3.2 crRNAs Designed by AutoCORDSv2 Were Sensitive for HTNV and SEOV Detection When Combined With RT-PCR for Preamplification
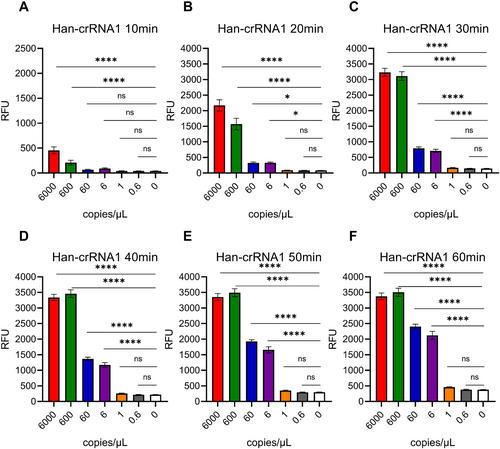
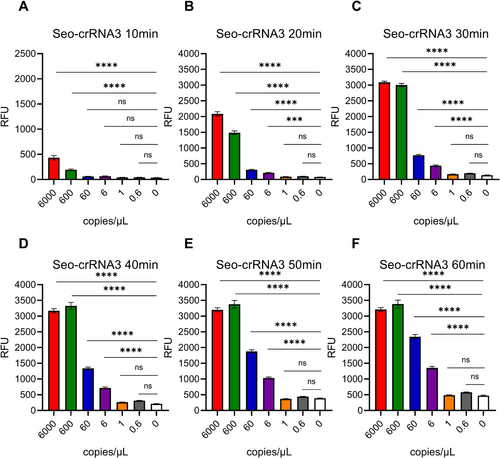
As most Cas12a-based nucleic acid detection assay, preamplification is important for enhancing the sensitivity of detection before Cas12a sensing reaction [11, 12, 16, 29-31]. There, we introduced RT-PCR to test the sensitivity of Han-crRNA1 and Seo-crRNA3, which was designed by AutoCORDSv2. First, target RNAs of HTNV and SEOV were transcribed in vitro by T7 RNA polymerase and quantified. Subsquently, HTNV and SEOV target RNAs were diluted as 6000, 600, 60, 6, 1, 0.6, and 0 copies/μL. RT-PCR was introduced to preamplification of target amplicons for Cas12a detection. Results indicated that a sensitivity of 6 copies/μL (equivalent to 111 copies per amplification reaction) could be achieved using Cas12a/Han-crRNA1 for the detection of HTNV with a reaction time of just 20 min (Figure 3 and Supporting Information S1: Figure S2A). Similarly, the sensitivity assay for Cas12a/Seo-crRNA3 in detecting SEOV also reached 6 copies/μL after a 20-min Cas12a reaction (Figure 4 and Figure S2B). The sensitivity of both Han-crRNA1 and Seo-crRNA3 is comparable to that of many Cas12-based detection methods, such as 10 copies/μL in Jiang 2023 [32] and Yang 2023 [33], 100 copies/μL in Wang 2023 [34], and 250 copies/μL in Tan 2024 [35]. The two crRNAs demonstrated ultrasensitive detection capabilities for HTNV and SEOV target RNAs, indicating that crRNAs designed by AutoCORDSv2 can be effectively utilized for the detection of hantaviruses and other pathogenic microorganisms.
3.3 Visual and Sensitive Detection of HTNV and SEOV Using RT-PCR-Cas12a
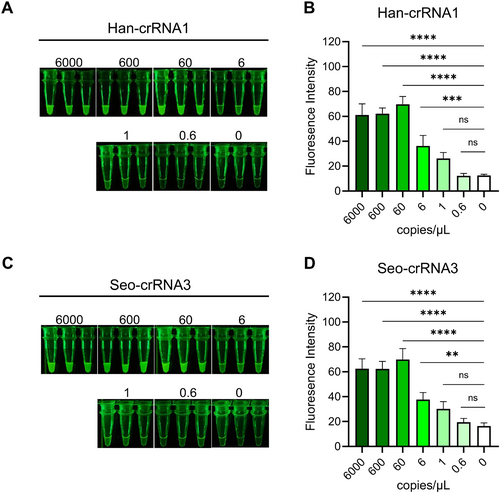
The visualization of results is more efficient and direct for on-site detection. The cleaved reporter, labeled with the FAM radical, can be visualized using a general blue light gel cutter, allowing for significant green light to be observed with the naked eye (Figure 1). In this study, we also evaluated the efficacy of two crRNAs (Han-crRNA1 and Seo-crRNA3) in visual detection. Following RT-PCR for preamplification and a 1-h Cas12a reaction, the results indicated that significant green light was present in tubes containing target amplicons compared to the blank control (Figure 5). For Han-crRNA1, the fluorescence intensity exhibited a gradient change corresponding to the target concentration gradient, with significant fluorescence detectable at a concentration as low as 6 copies/μL (equivalent to 111 copies per amplification reaction) (Figure 5A,B). In the case of Seo-crRNA3, the fluorescence was slightly higher than the control when only 6 copies/μL of the target was present, and this was evident when analyzing fluorescence intensity using ImageJ (Figure 5C,D). In conclusion, the crRNAs designed by AutoCORDSv2 can also be utilized for visual detection, demonstrating comparable sensitivity to results obtained using a microplate reader. Therefore, both Han-crRNA1 and Seo-crRNA3 are suitable for the visual and sensitive detection of HTNV and SEOV using the RT-PCR-Cas12a method.
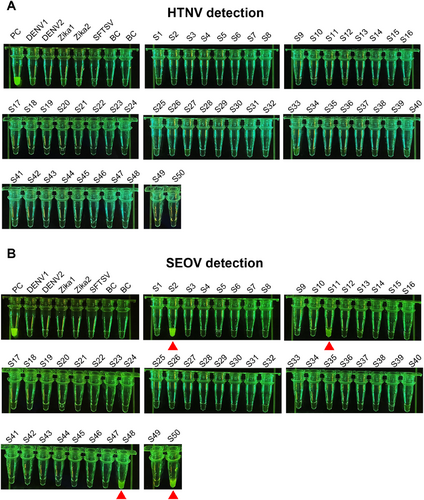
Next, 50 RNA samples obtained from the lungs of mice were analyzed using RT-PCR-Cas12a. Additionally, two samples of Dengue virus (DENV1 and DENV2), two samples of Zika virus (Zika1 and Zika2), and one sample of SFTSV were also tested to assess the specificity of Cas12a-Han-crRNA1 and Cas12a-Seo-crRNA3. In the detection of HTNV using Cas12a-Han-crRNA, all samples, with the exception of the positive control, did not exhibit an increase in fluorescence, indicating absence of HTNV infection in all 50 samples (Figure 6A). In the detection of SEOV, aside from the positive control, four samples (S1, S11, S48, and S50) demonstrated a significant increase in fluorescence (Figure 6B). The RT-qPCR results for these four samples were 25.58, 29.12, 26.95, and 26.80, which corresponded with the results from RT-PCR-Cas12a (Supporting Information S1: Figure S3). Furthermore, DENV, Zika virus, and SFTSV did not cause an increase in fluorescence, confirming that Han-crRNA1 and Seo-crRNA3 are specific for Hantavirus detection (Figure 6). These results collectively demonstrate the efficacy of Han-crRNA1 and Seo-crRNA3 for the visual and sensitive detection of real samples.
3.4 One-Pot RT-RPA-Cas12a Detection for HTNV and SEOV
To streamline the process of Hantavirus detection utilizing Cas12a-Han-crRNA1 and Cas12a-crRNA3, a one-pot RT-RPA-Cas12a system for the detection of HTNV and SEOV was developed (Figure 7A). This system integrates both the RT-RPA and the Cas12a/crRNA reactions within a single tube. In brief, sample RNA was introduced to the bottom of the tube containing the primers and RT-RPA components. Following a 30-min preamplification reaction, the Cas12a/crRNA reaction components, which were positioned in the lid of the tube, were mixed with the RT-RPA product through a short spin, initiating the Cas12a reaction. Several RT-RPA primers, which were automatically designed using AutoCORDSv2, were evaluated, leading to the selection of the efficient primer pairs H1 and S1 for further experimentation (Supporting Information S1: Figure S4 and Table S1). And results demonstrated that the presence of either HTNV or SEOV targets in the tube resulted in significant fluorescence, which was observable to the naked eye or via GelView under UV or blue light (Figure 7B). Subsequently, a SEOV RNA sample was analyzed using the one-pot RT-RPA Cas12a-Han-crRNA1 or Cas12a-Seo-crRNA3. Notably, fluorescence was only observed in the SEOV detection, indicating an increase in signal (Figure 7C,D).
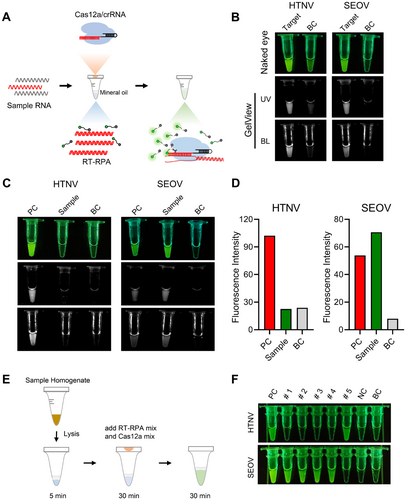
We further assessed the sensitivity of the one-pot assay, which demonstrated a sensitivity of 6 copies/μL (equivalent to 42 copies per amplification reaction), comparable to that of RT-qPCR (Supporting Information S1: Figure S5). Furthermore, the specificity of the assay was validated using dengue virus (DENV), Zika virus (ZIKV), and severe fever with thrombocytopenia syndrome virus (SFTSV), showing no cross-reactivity with other viruses (Supporting Information S1: Figure S6). Additionally, results from 50 RNA samples were fully concordant with commercial RT-qPCR assays (Supporting Information S1: Figures S3 and S6). To minimize assay steps, a 5-min sample lysis using acid treatment was combined with the one-pot RT-RPA-Cas12a detection (Figure 7E). Using this method, four SEOV samples and one HTNV sample were tested, and the results demonstrated that the method was both accurate and feasible (Figure 7F). This approach combines a rapid sample lysis technique with one-pot RT-RPA-Cas12a technology to achieve fast detection from sample to result. All these findings suggest that both Han-crRNA1 and Seo-crRNA3 can be effectively employed in the detection of HTNV and SEOV using the one-pot RT-RPA-Cas12a detection system.
4 Discussion
CRISPR detection has increasingly assumed a pivotal role in the field of diagnostics. During the COVID-19 pandemic from 2019 to 2022, numerous detection methods based on CRISPR/Cas12a were developed and implemented [34, 36-38]. In addition, methods utilizing CRISPR/Cas12a for the detection of enzymes, macromolecules, and antibodies have also been reported [39, 40]. However, there remains a deficiency in the automated integrated design of CRISPR/Cas12a system. Currently, the design of CRISPR RNA (crRNA) is predominantly reliant on online platforms, such as CHOPCHOP, Cas-OFFinder, and CRISPOR. These platforms generally design crRNA based on a single reference nucleic acid sequence and do not provide a conservation score for the sequence. In this context, our previously developed software, AutoCORDSv2, allows for the input of multiple nucleic acid sequences to calculate conservation scores. Designing crRNA based on conserved sequences results in crRNA that is more precise and specific. Furthermore, AutoCORDSv2 can return primers for specific amplification of the target sequence at both ends of the crRNA target, thereby facilitating the automated integrated design of the CRISPR/Cas12a detection system.
The crRNAs and primers auto-designed by AutoCORDSv2 exhibit high sensitivity and specificity. The two crRNAs, Han-crRNA1 and Seo-crRNA3, utilized for the detection of HTNV and SEOV viruses, demonstrate a sensitivity of up to 6 copies/μL (equivalent to 111 copies per reaction) when assessed through both direct visual observation and fluorescence reading devices. This sensitivity is comparable to, and in some cases exceeds, that of most existing CRISPR detection methods [33-36]. Furthermore, the sensitivity of these crRNAs is similar to that 10 copies/μL with RT-qPCR methods, or 3 copies/μL with RT-LAMP method [5, 41, 42]. In specificity experiments, no cross-reactivity was observed with three other viruses. Therefore, the crRNAs and primers designed by AutoCORDSv2, along with the established CRISPR/Cas12a detection system, provide a sensitivity comparable to RT-qPCR detection while allowing for direct visual observation of results.
The crRNA and primers designed by AutoCORDSv2 demonstrate a high degree of accuracy. The results obtained were consistent with those from a standardized RT-qPCR kit, suggesting that the accuracy of the CRISPR/Cas12a detection method utilized in this study is comparable to that of the standardized kit. Furthermore, the findings indicate that among the 50 nucleic acid samples from mouse lungs tested, only positive samples were detected in the SEOV detection. This observation is consistent with research conducted over the past decade, which indicates that the predominant hantavirus circulating in China is SEOV, rather than HTNV [43]. Future studies may focus on the selection of a single crRNA for hantavirus detection, contingent upon the prevalence of either HTNV or SEOV in different provinces.
The one-pot CRISPR detection method presents significant advantages for potential market applications. In recent years, one-pot CRISPR detection techniques have become a central focus of research. Several methodologies have been developed that integrate CRISPR/Cas12a detection with isothermal amplification, utilizing physical spatial separation to facilitate both preamplification and detection within a single tube [44-46]. Moreover, researchers have investigated light-controlled methods to modulate the activity of CRISPR/Cas12a detection, thereby minimizing the effects of Cas12a activation on amplification and promoting single-tube CRISPR detection [47, 48]. Additionally, studies have examined the design of crRNA to select suboptimal PAMs, which reduces the cis-cleavage efficiency of Cas12a and enhances the accumulation of target amplicons in a single-tube format [49, 50]. Among these one-pot Cas12a methodologies, we adopt the most widely utilized approach, wherein Cas12a/Han-crRNA1 and Cas12a/Seo-crRNA3 are positioned in the tube cap while RT-RPA occurs at the bottom of the tube. This configuration enables single-tube detection of hantavirus, thereby further simplify the hantavirus detection process. The sensitivity of one-pot RT-RPA-Cas12a is 6 copies/μL (equivalent to 42 copies per reaction), which is comparable to the 10 copies/μL sensitivity achieved using RT-qPCR [40, 41]. The specificity and accuracy of sample detection were fully concordant with those of commercial RT-qPCR assays. Furthermore, a rapid sample lysis technique was integrated with one-pot RT-RPA-Cas12a technology to facilitate fast detection from sample to result. This study also demonstrates that the crRNAs and primers designed by AutoCORDSv2 are suitable for implementation in a single-tube CRISPR/Cas12a detection system.
5 Conclusion
In this study, we developed a CRISPR/Cas12a-based method for the detection of hantaviruses utilizing AutoCORDSv2. By integrating RT-PCR technology, we achieved sensitive, specific, and visual detection of Hantaan virus and Seoul virus. The results obtained from the detection of RNA samples were consistent with those obtained from commercially available RT-qPCR kits. Furthermore, by incorporating RT-RPA technology, we successfully implemented a one-tube detection system, which minimized the risk of cross-contamination and enhanced operational simplicity. Our findings further suggest that AutoCORDSv2 has significant potential for the rapid and automated design of Cas12a/crRNA detection systems for various pathogens.
Author Contributions
Changsheng He: writing – original daft, designed and performed experiments and wrote the manuscript. Wei Zhu: writing – original daft, designed and performed experiments and wrote the manuscript. Xing Zhang: performed variant RNA samples assay. Wei Wu: performed variant RNA samples assay. Jiexing Tan: performed variant RNA samples assay. Bosheng Li: advised on experimental design. Dongchao Huang: formal analysis, performed bioinformatic analysis of sequences. Yinghua Chen: formal analysis, analyzed the data. Zhiguang Xiang: advised on experimental design. Lizhen Huang: supervision, conceived and designed the study, supervised the study. Li Gong: supervision, writing – original daft, funding acquisition, conceived and designed the study, supervised the study, wrote manuscript and acquired the research funds.
Acknowledgments
This study is financially supported by The President Foundation of Nanfang Hospital, Southern Medical University (grant number 2023B036); National Science and Technology Infrastructure of China (Project No. National Pathogen Resource Center-NPRC-32); The CAMS Innovation Fund for Medical Sciences (grant number 2021-I2M-1-039); National Key R&D Program of China, grant number (grant number 2022YFC2303400); The Guangdong Basic and Applied Basic Research Foundation (grant number 2023A1515140080); Dongguan Social development technology projects Science and Technology Planning Project (grant number 20231800940792).
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
All data pertinent to this study are presented in the main text. The data that underpin the findings of this study are accessible in the supporting materials accompanying this article.




