Metagenomic analysis of the gut virome in patients with irritable bowel syndrome
Abstract
Irritable bowel syndrome (IBS), a chronic functional gastrointestinal disorder, is recognized for its association with alterations in the gut microbiome and metabolome. This study delves into the largely unexplored domain of the gut virome in IBS patients. We conducted a comprehensive analysis of the fecal metagenomic data set from 277 IBS patients and 84 healthy controls to characterize the gut viral community. Our findings revealed a distinct gut virome in IBS patients compared to healthy individuals, marked by significant variances in between-sample diversity and altered abundances of 127 viral operational taxonomic units (vOTUs). Specifically, 111 vOTUs, predominantly belonging to crAss-like, Siphoviridae, Myoviridae, and Quimbyviridae families, were more abundant in IBS patients, whereas the healthy control group exhibited enrichment of 16 vOTUs from multiple families. We also investigated the interplay between the gut virome and bacteriome, identifying a correlation between IBS-enriched bacteria like Klebsiella pneumoniae, Fusobacterium varium, and Ruminococcus gnavus, and the IBS-associated vOTUs. Furthermore, we assessed the potential of gut viral signatures in predicting IBS, achieving a notable area under the receiver operator characteristic curve (AUC) of 0.834. These findings highlight significant shifts in the viral diversity, taxonomic distribution, and functional composition of the gut virome in IBS patients, suggesting the potential role of the gut virome in IBS pathogenesis and opening new avenues for diagnostic and therapeutic strategies targeting the gut virome in IBS management.
1 INTRODUCTION
Irritable bowel syndrome (IBS) is a functional bowel disease characterized by abdominal pain, distension, or discomfort associated with altered bowel habits,1, 2 affecting approximately 5%–10% of the general population and significantly impacting the quality of life and social functioning.3 The pathophysiology of IBS remains poorly understood, with potential risk factors including genetic predisposition, chronic infection, microbiota dysregulation, immune activation, low-grade mucosal inflammation, altered intestinal permeability, bile salt metabolism disorders, abnormal serotonin metabolism, and altered brain function.4
Treatment strategies for IBS aim to alleviate abdominal pain and improve bowel habits through dietary modifications, soluble fiber intake, anti-spasmodic drugs, central neuromodulators, antibiotics, and psychotherapy for severe cases.5 Recent evidence highlights the involvement of gut microbiota in IBS development, with dysbiosis and interactions with the gut-brain axis implicated in pathogenesis.6 Moreover, previous gastrointestinal infections have been linked to IBS, supporting the role of microbiota dysbiosis in the disease.7 Next-generation sequencing has revealed a significantly lower abundance of intestinal Lactobacilli, Bifidobacterium, and Faecalibacterium prausnitzii in IBS patients compared to healthy controls.8 Fecal microbiota transplantation (FMT) was able to alter the gut microbial composition in IBS patients, improving symptoms and quality of life in patients9; it also demonstrated the ability to optimize intestinal barrier function and host immunity in animal models.10 Importantly, different IBS clinical subtypes (such as constipation and diarrhea) and severities exhibit distinct characteristics in the gut microbiome,11-13 suggesting a heterogeneous connection between the gut microbiota and disease pathogenesis. Besides, gut microbiota-derived metabolites, such as bile acids, short-chain fatty acids (SCFAs), vitamins, amino acids, serotonin, and hypoxanthine, are also closely associated with IBS pathogenesis.14
Given that the gut microbiota contains various components, including bacteria, fungi, and viruses, a notable gap in the previous studies is the limited exploration of the distinct roles each component plays. While current research predominantly focuses on gut bacteria, the significance of gut viruses should not be overlooked. The global virome database indicates that 97.7% of human viral populations consist of bacterial viruses (phages), with 2.1% being eukaryotic viruses and 0.1% archaeal viruses, many of which are uncultured and/or unclassified.15 The gut virome closely interacts with the microbiota, and recent studies underscore its association with various diseases, particularly inflammatory bowel disease (IBD), obesity, diabetes, stress, and autoimmune and liver diseases.16-21 It is crucial to comprehend the diversity and structural changes of the gut virome during disease to explore the underlying pathogenesis. Ansari et al. investigated the fecal virome of 25 IBS patients and 17 controls using the subset of reads that can be classified into known viral families by alignment to the Viral RefSeq database.22 Another metagenomic sequencing of fecal virus-like particles (VLP) from 55 IBS patients and 51 control individuals revealed significantly lower α-diversity of viral clusters in the IBS groups and a distinct β-diversity compared to the healthy controls, although not among different IBS symptom subtypes.23 They also identified the core virome consisting of 5 and 12 viral clusters in IBS and control subjects, respectively.23
In this context, we conducted a comprehensive study to examine both known and unknown viral sequences in IBS patients. The analysis flowchart is shown in Figure S1. Through the reanalysis of publicly available metagenomic data from a cohort of 361 human fecal samples, we aimed to identify changes in the gut virome associated with IBS. Specifically, we compared the viral composition between IBS patients and healthy individuals based on metagenomic sequencing data and explored the intricate relationship between viruses and bacteria. Our goal is to gain a deeper understanding of the etiology and pathogenesis of IBS through the investigation of gut viruses in IBS patients. Such insights may pave the way for new perspectives on prevention and treatment strategies for IBS.
2 MATERIALS AND METHODS
2.1 Subjects and datasets
This study analyzed fecal samples from a total of 361 subjects, comprising 277 individuals diagnosed with IBS and 84 healthy individuals of China origin. The data set was sourced from a study conducted by Han et al.,24 and the publicly available fecal metagenomic samples was retrieved from the China National GeneBank (CNGB) Nucleotide Sequence Archive, with the accession number CNP0000334. To ensure comparability, the patients and healthy controls were matched based on their gender (% female in patients vs. controls: 56% vs. 68%), age (patients ranged from 18 to 64 [mean, 43.8], controls ranged from 19 to 64 [mean, 40.3]), and body mass index (BMI), utilizing phenotype information from the original study. To ensure data quality, raw metagenomic reads underwent a rigorous quality control process using fastp.25 Reads exhibiting low quality (defined as having more than 50% bases with a quality score below 20 or more than 5 “N” bases), low complexity, or contained adapter sequences were excluded. The remaining reads were trimmed at the tails to eliminate low-quality (<Q20) or “N” bases. Additionally, human genomic sequences were removed from the high-quality metagenomic reads by aligning them with the reference human genome (GRCh38) using Bowtie2.26 To validate the efficiency of IBS viral signatures in an independent cohort, we included and analyzed a fecal metagenomic data set from Tap et al.'s study, consisting of 104 IBS patients and 38 healthy controls of Sweden origin.12 This data set was downloaded from the EMBL European Nucleotide Archive under accession number PRJEB34992. Metagenomic reads were processed using the same approaches employed in the Chinese cohort.
2.2 Gut virome profiling and analyses
To analyze the gut virome of the fecal metagenomic data set, we employed the Chinese gut virus catalog (cnGVC), which contained over 67 000 nonredundant viral operational taxonomic units (vOTUs), as a reference. The cnGVC was constructed from a comprehensive data set of more than 10 000 publicly available fecal metagenomes from the Chinese population, including all samples used in our current study.27 Briefly, the clean reads of each metagenomic sample were assembled into contigs using Megahit v1.2.9 with the parameters “--k-list 21,41,61,81,101,121,141.”28 Contigs with a length of ≥5 kb were utilized for identifying viral sequences in each sample. Subsequently, identification, decontamination, and dereplication of viral sequences were performed following previously established methods.29-31 The quality of vOTUs was assessed using CheckV v0.7.0.32 Taxonomic classification, host prediction, and functional annotation of viral sequences were also performed according to the prior study.29
To elucidate the taxonomic profiles of the gut viral community, the clean reads from each fecal metagenomic sample were mapped to the reference genomes of all cnGVC vOTUs using Bowtie2 v2.4.1 with the parameters “--end-to-end --fast --no-unal” (Figure S1). To ensure comparability, the total mapping reads of all samples were randomly subsampled to the same sequencing amount (4 million reads). The relative abundance of each vOTU was calculated as the number of reads mapped to that vOTU divided by the total mapping reads. Additionally, the relative abundance of each viral family was obtained by summing the relative abundances of vOTUs annotated with that specific family.
For assessing the gut virome diversity, we analyzed the profiles at the vOTU level. The total number of observed vOTUs was determined by counting the number of vOTUs with a relative abundance greater than zero. To further quantify the diversity of the gut virome, Shannon and Simpson diversity indices were calculated using the diversity function of the R vegan package.
2.3 Functional annotation of the viral genomes
We performed functional annotation of the vOTUs using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database.33 To achieve this, the viral protein-coding genes for each vOTU were aligned against the KEGG database using DIAMOND 34 with the following options “-query-cover 50 -subject-cover 50-e 1e-5 -min-score 50 -max-target-seqs. 50.” Then, each protein was then assigned a KEGG ortholog (KO) based on the best-hit protein in the database. Moreover, we identified the viral auxiliary metabolic genes (AMGs) within the vOTUs following the methodology described in a previous study.35
2.4 Gut bacteriome profiling
We conducted bacterial taxonomic profiling at various levels, including phylum, class, order, family, genus, and species levels, on the fecal metagenomic data set for both IBS patients and healthy controls using MetaPhlAn 3.36 This tool utilizes clade-specific marker genes to accurately classify metagenomic reads to their respective taxonomies and generate relative abundances of identified taxa in the samples. To calculate the relative abundance of each bacterial species, we randomly selected 10 million reads from each fecal sample.
2.5 Statistical analyses and visualization
The statistical analyses were conducted using the R v4.0.1 platform and included various methods such as distance-based redundancy analysis (dbRDA), principal coordinates analysis (PCoA), permutational multivariate analysis of variance (PERMANOVA), Student's t-test, Wilcoxon rank-sum test, Spearman's correlation analysis, and random forest analysis. dbRDA and PCoA were performed based on the Bray-Curtis distance and visualized using the vegan package.37 PERMANOVA was executed using the adonis function from the vegan package, and the adonis p-value was generated with 1000 permutations. The Student's t-test and Wilcoxon rank-sum test were used to measure statistical differences in the diversity and taxonomic levels, respectively, between the two cohorts. The q-values were used for multiple testing correction and generated by the Benjamini-Hochberg procedure. A p-value (for a single test) or q-value (for multiple testing) less than 0.05 was considered statistically significant.
Spearman's correlation analysis was used to quantify the associations between viruses and bacteria. Correlations with an absolute correlation coefficient >0.50 and a q-value < 0.05 were represented in the correlation network. For each virus-bacterium pair, a correlation coefficient was computed based on the relative abundances, while adjusting for individuals' gender, age, and body mass index (BMI). The correlation network was visualized using Cytoscape.38
Random forest models, comprising 1000 trees, were trained with the randomForest package to discern between IBS patients and healthy controls. For within-cohort analysis, we initially trained a random forest model based on the abundance profiles of the differential viral signatures and used leave-one-out cross-validation to predict the status of each subject based on the model. For between-cohort analysis, we trained a random forest model based on the abundance of the IBS-associated signatures in the Chinese cohort and tested its performance in distinguishing IBS patients and healthy controls in the Swedish cohort. The model's performance was assessed by calculating the area under the receiver operating characteristic curve (AUC) using the roc function. The importance ordering of markers was determined via the importance function.
3 RESULTS
3.1 Data summary and gut viral diversity
A total of 1.85 Tbp of fecal metagenomic sequencing data, with an average of 5.12 ± 1.2 Gbp per fecal sample, was obtained from a previous study involving 277 IBS patients and 84 healthy subjects.24 The raw metagenomic data set was quality controlled and decontaminated for the human sequences, generating a total of 1.84 Tbp of high-quality data (average 5.09 ± 1.2 Gbp per sample) for all subjects. Then, the high-quality reads were mapped into the cnGVC catalog constructed from Chinese populations (comprising 67 096 vOTUs; see “Materials and methods”), and a profile of the gut viral composition of all fecal samples was obtained for subsequent analysis.
Firstly, we examined the within-sample viral richness and diversity between IBS patients and healthy subjects. At the vOTU level, the viral richness, estimated by the observed number of vOTUs, showed a slightly higher trend in IBS patients compared with healthy controls, although not statistically significant (Student's t-test p = 0.063). However, viral diversity, assessed by the Shannon and Simpson indices, exhibited no significant difference between the two groups (Figure 1A). Likewise, at the family level, both viral richness and diversity did not demonstrate significant disparities (Figure 1B). Furthermore, we investigated the between-sample viral diversity between IBS patients and healthy controls using the Bray-Curtis distance metric. The results revealed that the gut virome of IBS patients exhibited a higher average pairwise distance compared to that of healthy subjects (Student's t-test p < 0.001 at the vOTU and family levels; Figure 1C). Additionally, the average pairwise distance between IBS patients and healthy controls was higher than the average distance among healthy control subjects but lower than the average distance among IBS patients.
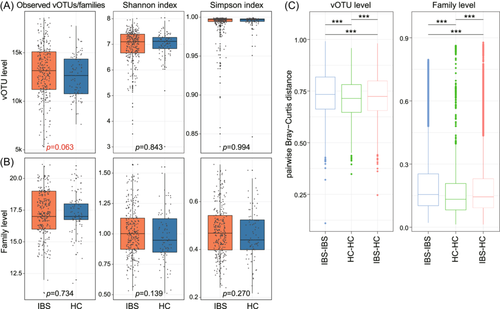
3.2 Structure changes of the gut virome in IBS patients
Next, we conducted a dbRDA analysis using the Bray-Curtis distance to investigate the distinctions in the gut virome between IBS patients and healthy subjects. This analysis revealed that the samples from the two groups were clearly separated (PERMANOVA p = 0.011; Figure 2A), suggesting considerable structural changes in the gut virome of IBS patients. At the family level, the viromes of both patients and controls exhibited dominance of Siphoviridae, Myoviridae, Quimbyviridae, crAss-like members, and a substantial proportion of viruses belonging to unclassified taxa (Figure 2B). Notably, the relative abundances of crAss-like, along with several low-abundance families such as Metaviridae, Mitoviridae, and Circoviridae, were significantly reduced in the virome of IBS patients compared with that in healthy controls (Wilcoxon rank-sum test p < 0.05; Figure 2C; Table S1). Conversely, two families, Podoviridae and Caulimoviridae, showed significant enrichment in IBS patients.
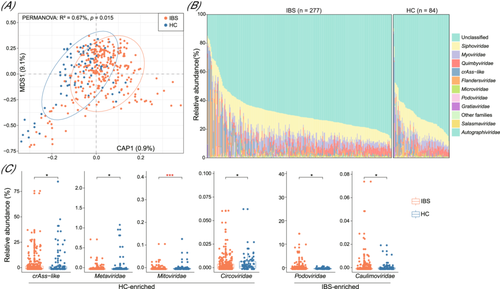
3.3 Gut viral signatures associated with IBS
Next, we compared the viral profiles between patients and controls at the vOTU level to identify gut viral signatures associated with IBS. We focused on 11 588 core vOTUs with an average relative abundance greater than 0.0005% and an occurrence rate exceeding 20% in all samples, representing an average of 93.3% of the total virome abundances in the samples. Among these, 127 vOTUs exhibited significant differences in relative abundances between the two groups (Wilcoxon rank-sum test q < 0.05 and |fold-change| > 2; Figure 3A; Figure S2; Table S2) with 111 vOTUs enriched in the virome of IBS patients and 16 vOTUs enriched in those of healthy individuals. The majority of the IBS-enriched vOTUs belonged to unclassified (n = 31), crAss-like (n = 30), and Siphoviridae viruses (n = 21), followed by Myoviridae (n = 14), Quimbyviridae (n = 8), and several low-abundance viral families including Flandersviridae, Gratiaviridae, and Podoviridae (Figure 3B). Inversely, the control-enriched vOTUs comprised 5 crAss-like, 3 unclassified, 2 Myoviridae, 2 Flandersviridae, 1 Siphoviridae, 1 Quimbyviridae, 1 Gratiaviridae, and 1 Microviridae viruses. Additionally, prokaryotic host assignment of the vOTUs indicated that a considerable number of IBS-enriched vOTUs could be predicted to infect Fusobacteriaceae (n = 11), Lachnospiraceae (n = 7), Enterobacteriaceae (n = 6), and Bacteroidaceae (n = 5) bacteria (Figure 3B; Table S2).
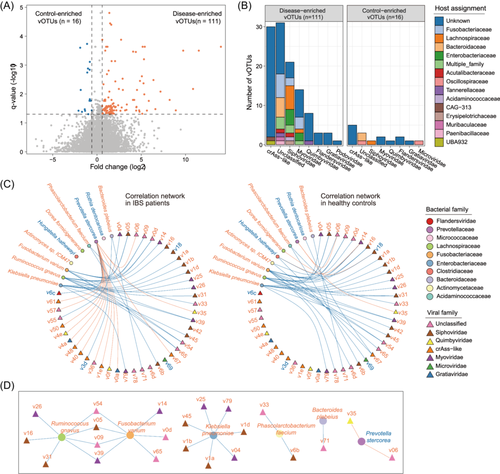
To uncover the connections between the IBS-associated gut viral signatures and the gut bacteria, we initially profiled the gut bacteriome of all samples and identified 33 bacterial species (11 IBS-enriched and 22 control-enriched) showing significant differences between IBS patients and healthy individuals (Wilcoxon rank-sum test q < 0.05; Table S3). Subsequently, employing Spearman correlation coefficient analysis, we uncovered 43 strong virus-bacterium correlations within the IBS patient samples and 38 correlations within the healthy subjects (absolute correlation coefficient ρ > 0.50; Figure 3C; Table S4). Notably, 25 of these correlations were shared between the two cohorts. Certain IBS-enriched bacterial species, including Klebsiella pneumoniae, Fusobacterium varium, and Ruminococcus gnavus, demonstrated positive correlations with the highest number of IBS-enriched vOTUs, while an IBS-depleted species, Prevotella stercorea, exhibited negative correlations with two control-enriched vOTUs (Figure 3D; Table S4). These results suggest potential collaborative interactions between these gut bacteria and viruses in influencing the disease.
3.4 Functions of the IBS-associated viruses
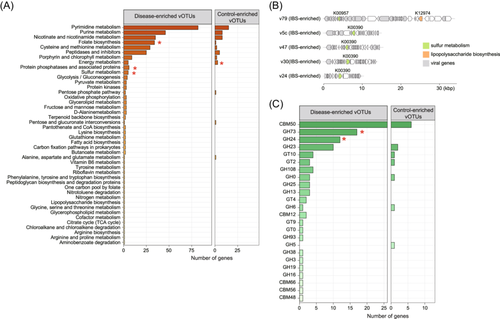
To gain insights into the functional characteristics of the gut viral signatures, we predicted a total of 15 952 genes from the 127 IBS-associated vOTUs and functionally annotated 8.8% (1397/15 952) of these genes based on the KEGG database (Figure S2). Over one-fourth (27.2%) of the annotated genes, corresponding to 2.4% of all genes, were further identified as viral AMGs following a previously established method (see Materials and methods). Notably, functions related to nucleotide and amino acid metabolism, as well as other compounds, were among the most commonly encoded AMGs. At the KEGG pathway level C, functions involved in folate biosynthesis, protein phosphatases and associated proteins, and sulfur metabolism were more prevalent in the IBD-enriched vOTUs when compared with the control-enriched vOTUs, whereas functions associated with energy metabolism were more abundant in the control-enriched vOTUs (Fisher's exact test, q < 0.05; Figure 4A). Of particular interest were two enzymes, phosphoadenosine phosphosulfate reductase (K00390) and sulfate adenylyltransferase (K00957), involved in assimilatory sulfate reduction, which were notably present in the disease-enriched vOTUs and could be linked to mechanisms in IBS patients (Figure 4B). Additionally, we identified 113 auxiliary carbohydrate-active enzymes (CAZymes), corresponding to 0.7% of all genes, from the IBS-associated viruses. Notably, two types of CAZymes, glycoside hydrolase family 73 (GH73) and glycoside hydrolase family 24 (GH24), were significantly more abundant in IBD-enriched vOTUs compared with control-enriched vOTUs (Figure 4C). These findings provide valuable insights into the potential functional roles of the gut viral signatures associated with IBS.
3.5 Prediction of IBS state using gut viral signatures
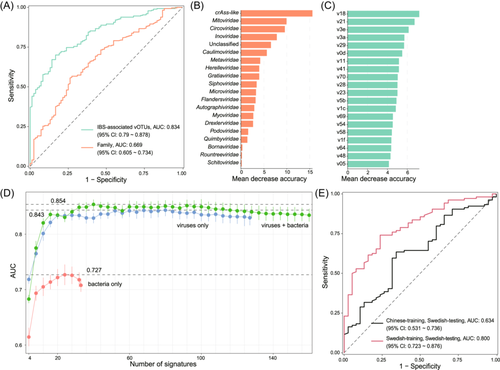
Finally, we assessed the predictive ability of gut viral signatures in identifying the IBS state. By training a random forest classifier based on the relative abundances of the 127 IBS-associated vOTUs, we achieved an impressive AUC of 0.834 (95% confidence interval [CI], 0.790–0.878) for distinguishing between IBS patients and healthy controls (Figure 5A). Additionally, a classifier trained using the relative abundances of viral families yielded an AUC of 0.669 (95% CI, 0.605–0.734). Several viral families and vOTUs displayed the highest scores in the classifiers (Figure 5B–C; Table S5), indicating their potential diagnostic significance for the disease. Furthermore, to generate a minimal set of gut microbial signatures for accurate IBS classification, we trained new random forest models using the most important viruses and bacteria. The viral model demonstrated the optimal AUC of 0.843 when employing a subset of the top 80 important vOTUs (Table S6), highlighting the strong diagnostic potential of gut viral signatures in predicting the IBS state. As a comparison, the virus-bacterium combined model achieved the highest AUC of 0.854 with 40 viruses/bacteria while the model using only bacterial signatures generated the highest AUC of 0.727 with 24 bacteria (Figure 5D; Table S6). To further validate the utility of gut viral signatures in distinguishing the IBS state, we introduced a new data set obtained from a gut metagenome study involving 104 IBS patients and 38 healthy controls of Swedish origin and profiled the composition of 127 IBS-associated viruses (i.e., 111 IBS-enriched and 16 control-enriched vOTUs) for these samples. When constructing a random forest model and performing leave-one-out training and prediction with these Swedish samples, we achieved an optimal AUC of 0.800 (95% CI, 0.723–0.876) (Figure 5E). However, when using the original Chinese samples to build a random forest model and predict on the Swedish samples, the AUC reached only 0.634 (95% CI, 0.531–0.736). These results suggest that the IBS-associated viruses we identified exhibit a good discriminatory effect between IBS and healthy states, but this discrimination does not perform as well across different populations, possibly due to variations in different populations or datasets.
4 DISCUSSION
Irritable bowel syndrome is recognized as the most prevalent functional gastrointestinal disorder.3 Characterized by recurring abdominal pain and irregularities in stool form or frequency, IBS presents in three clinical subtypes: IBS with constipation (IBS-C), IBS with diarrhea (IBS-D), and IBS with mixed bowel habits (IBS-M).2 Previous studies have extensively explored alterations in the bacterial aspect of the gut microbiome in IBS patients, revealing dysbiosis compared to healthy individuals.39 However, the role of the gut virome remains largely overlooked. In our study, we investigated changes in the gut viral population in 277 IBS patients compared to 84 healthy controls, enhancing the understanding of the gut virome in IBS through metagenomic sequencing.
Our results revealed relatively stable within-sample diversity but disturbed between-sample diversity in the gut virome of IBS patients. Additionally, when comparing the family-level composition, we found that IBS patients had an increased abundance of Podoviridae and Caulimoviridae, while healthy controls showed a higher abundance of Metaviridae, Mitoviridae, and Circoviridae in their viromes. The enrichment of Podoviridae is in line with previous reports associating it with autoimmune diseases such as systemic lupus erythematosus (SLE).40 On the other hand, Caulimoviridae is a plant virus that has not been known to cause human disease, while Metaviridae is a retrovirus family primarily infecting fungi and invertebrates, and Mitoviridae typically exhibit symptomless infections in their host, including fungi.41 Circoviridae is widely found in both mammals and birds and has been previously detected in adult respiratory samples.42
At the vOTU level, IBS-enriched vOTUs belonged to crAss-like, Siphoviridae, Myoviridae, Quimbyviridae, as well as several less prevalent viral families like Flandersviridae and Gratiaviridae, while control-enriched vOTUs primarily consisted of crAss-like and some other viral families. CrAss-like viruses, known to be abundant in the human gut,43 have been associated with various diseases, including IBD,44, 45 RA, SLE,40 Parkinson's disease, atherosclerosis,46 liver cirrhosis, and ankylosing spondylitis.47 Siphoviridae, Myoviridae, and Podoviridae were also reported to be enriched in IBS patients in other gut virome studies,23 as well as in CRC patients,48 further supporting our findings. Quimbyviridae is a recently described novel family of viruses widely abundant in the human gut, primarily infecting Bacteroidetes.49 Gratiaviridae is another new viral family known to infect Bacteroides and is remotely related to several other viral families.49 Flandersviridae, a member of Caudovirales, is a common and abundant viral family found in metagenomes throughout the community.49 Microviridae phages, the most stable colonizers in the human gut, have been observed to be elevated in IBS-D and IBS-C,50 aligning with our study's outcomes.
In terms of the viral host aspect, several IBS-enriched bacterial species, such as Klebsiella pneumoniae, Fusobacterium varium, and Ruminococcus gnavus, exhibiting positive correlations with a substantial number of IBS-enriched vOTUs. Conversely, an IBS-depleted species, Prevotella stercorea, demonstrated negative connections with two control-enriched vOTUs. Previous research using 16 S rDNA amplicon sequencing and quantitative PCR (qPCR) analyses has also shown elevated levels of Klebsiella spp in IBS patients.51 Additionally, Klebsiella pneumoniae was found to be the most common isolate among patients with small intestinal bacterial overgrowth (SIBO),52 which is commonly found in most IBS patients, indicating the need for further attention to the relationship between Klebsiella pneumoniae and IBS. Fusobacterium varium is primarily found in the mucosa of humans and animals, with a significant presence in the mouth and colon. It has been closely associated with IBD 53 and detected in inflamed intestinal tissues of ulcerative colitis (UC) patients.54 Ruminococcus gnavus, which was found in the mucosal biofilms of IBS and UC patients, may play a pathogenic role in IBS-D by increasing serotonin biosynthesis.55, 56 Prevotella can be used as opportunistic pathogens, causing periodontal and dental, intestinal inflammation, rheumatoid arthritis, and bacterial vaginitis.57 Multiple studies have reported an increased abundance of Prevotella in IBS patients,58 and its prevalence decreased with the increase in IBS symptom severity.11 These findings highlight the distinct virus-host relationships between IBS-enriched and healthy control-enriched vOTUs.
Regarding the functions of viral signatures associated with IBS, we identified enzymes involved in assimilatory sulfate reduction, including phosphoadenosine phosphosulfate reductase K00390 and sulfate adenylyltransferase K00957, that were particularly prevalent in disease-enriched vOTUs. These enzymes might be linked to the mechanism underlying IBS in patients. Additionally, we identified 113 auxiliary carbohydrate-active enzymes (CAZymes) from the IBS-associated viruses. Two types of CAZymes, glycoside hydrolase family 73 (GH73) and glycoside hydrolase family 24 (GH24), were significantly more abundant in disease-enriched vOTUs compared to healthy control-enriched vOTUs.
Until now, the identification of biomarkers for IBS has been a topic of extensive research, with several potential candidates proposed. For instance, Priya et al. reported that bile and fat excretion could serve as biomarkers for clinically significant diarrhea and constipation in IBS, with AUC values of >0.82 and >0.71, respectively.59 Another study highlighted the potential of the density of rectal peptide YY and somatostatin cells as biomarkers for the diagnosis of IBS, achieving high AUC values of 0.99 and 0.86, respectively.60 In our study, we assessed the diagnostic potential of the gut virome in predicting IBS using the relative abundances of 127 IBS-associated vOTUs and obtained an AUC value of 0.834. This result suggests that the gut virome has significant predictive power in discriminating IBS cases from healthy controls. Furthermore, we tested the utility of these 127 IBD-associated vOTUs in predicting IBS within a Swedish cohort, achieving a within-cohort AUC of 0.800. However, when applied to a between-cohort scenario (Chinese cohort training and Swedish cohort testing), the AUC was lower at 0.634. These findings are encouraging and indicate that the gut virome can be a valuable source of potential biomarkers for IBS, adding to the growing body of evidence supporting its high diagnostic potential, nonetheless, cross-population predictions require further investigation.
Despite these promising results, our study has several limitations. First, while we considered individual phenotypes such as gender, age, and BMI in the analysis, other confounding factors, including diet, medication, lifestyle, and environmental exposure, were not considered due to the lack of information. Many studies have demonstrated the significant impact of environmental factors on the gut microbiota,61-64 and specifically, individual lifestyle factors such as diet and physical activity have been shown to have strong associations with the gut virome.65, 66 Second, due to the relatively small sample size in the current study, we did not comprehensively stratify IBS patients into disease subtypes or severity levels. Future large-scale studies are warranted to investigate the virome characteristics of different IBS patient subgroups. Additionally, due to the limitations in the experimental design, we cannot conclusively determine whether changes in the gut virome of IBS patients are causative or consequential to the disease. Prospective longitudinal studies or mechanistic studies in animal models may provide insights in this regard.
5 CONCLUSION
In conclusion, our study provides valuable insights into the gut virome in IBS. We observed considerable structural changes in the gut virome of IBS patients compared to healthy controls, highlighting the potential role of viral populations in the disease's pathogenesis. The enrichment of specific viral families, such as Podoviridae and Caulimoviridae, in IBS patients suggests their potential as markers for disease association. Furthermore, our analysis of viral-host interactions identified several IBS-enriched bacterial species, such as Klebsiella pneumoniae, Fusobacterium varium, and Ruminococcus gnavus, showing positive correlations with IBS-enriched vOTUs, while Prevotella stercorea displayed negative associations with control-enriched vOTUs. In summary, our findings emphasize the significance of the gut virome in IBS and its potential as a valuable resource for identifying biomarkers and improving diagnostic approaches. Further research and validation studies in larger cohorts are needed to harness the full potential of the gut virome in understanding and managing IBS, potentially leading to more effective and personalized treatment strategies for individuals affected by this common functional gastrointestinal disorder.
AUTHOR CONTRIBUTIONS
Research Design: Pan Zhang, Haitao Shi, Ruochun Guo, Jinhai Wang, and Shiyang Ma were involved in the design of the study. Data Analysis and Discussion: Pan Zhang, Shenghui Li, Yue Zhang, Jiong Jiang, Ting Wang, Cui Fu, Huan Zhang and Shenghui Li carried out the data analysis and contributed to the discussion of the results. Drafting of the Manuscript: The initial draft of the manuscript was written by Pan Zhang, Haitao Shi, Ruochun Guo, Lu Li, Xiaoyan Guo, Danyan Chang, Longbao Yang and Shenghui Li. Revision of the Manuscript: The manuscript was revised by Pan Zhang, Haitao Shi, Ruochun Guo, Jinhai Wang, and Shiyang Ma. All authors read and approved the final manuscript.
ACKNOWLEDGMENTS
We extend our heartfelt thanks to all the staff of the Department of Gastroenterology at the Second Affiliated Hospital for their support and contributions to this research.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
ETHICS APPROVAL
Not applicable. This study involves secondary analysis of publicly available data, hence no ethical approval was required.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available in CNGB, NCBI at https://www.ncbi.nlm.nih.gov/,https://db.cngb.org/. These data were derived from the following resources available in the public domain: - PRJEB34992, https://www.ncbi.nlm.nih.gov/sra/?term=PRJEB34992-CNP0000334, https://db.cngb.org/search/?q=CNP0000334. The data supporting the findings of this study are available within the article. Further details about the data and how it can be accessed are outlined in the Methods section of the manuscript.




