First detection of multiple cases related to CV-A16 strain of B1c clade in Beijing in 2022
Abstract
Coxsackievirus A16 (CV-A16) is a significant etiologic agent of hand, foot, and mouth disease (HFMD) and herpangina (HA), with the capacity to progress to severe complications, including encephalitis, aseptic meningitis, acute flaccid paralysis, myocarditis, and other critical conditions. Beijing's epidemiological surveillance system, established in 2008, encompasses 29 hospitals and 16 district disease control centers. From 2019 to 2021, the circulation of CV-A16 was characterized by the co-circulation of B1a and B1b clades. Multiple cases of HFMD linked to clade B1c has not been reported in Beijing until 2022. This study enrolled 400 HFMD and 493 HA cases. Employing real-time RT-PCR, 368 enterovirus-positive cases were identified, with 180 selected for sequencing. CV-A16 was detected in 18.89% (34/180) of the cases, second only to CV-A6, identified in 63.33% (114/180). Full-length VP1 gene sequences were successfully amplified and sequenced in 22 cases, revealing the presence of clades B1a, B1b, and B1c in 14, 3, and 5 cases, respectively. A cluster of five B1c clade cases occurred between June 29 and July 17, 2022, within a 7-km diameter region in Shunyi District. Phylogenetic analysis of five complete VP1 gene sequences and two full-genome sequences revealed close clustering with the 2018 Indian strain (GenBank accession: MH780757.1) within the B1c India branch, with NCBI BLAST results showing over 98% similarity. Comparative sequence analysis identified three unique amino acid variations (P3S, V25A, and I235V). The 2022 Shunyi District HFMD cases represent the first instances of spatiotemporally correlated CV-A16 B1c clade infections in Beijing, underscoring the necessity for heightened surveillance of B1c clade CV-A16 in HFMD and HA in this region.
1 INTRODUCTION
Hand, foot, and mouth disease (HFMD) was class C notifiable disease in China. The clinical symptoms include fever and rash or herps in hand, foot, or mouth. Some patients may develop severe complications, i.e. myocarditis and aseptic meningoencephalitis. HFMD is common among children under 5 years old. The incubation period is 4–7 days and is highly contagious.1 Various enteroviruses (EVs) could be the causative agent of HFMD, and group A coxsackievirus 16 (CV-A16) is an important pathogen in recent years.2, 3 Another acute respiratory infectious disease caused by enteroviruses is herpangina (HA).4 The main clinical feature is painful oral ulcers in the posterior part of the oral cavity and fever. Comparing to HFMD, the occurrence of ulcers in HA is limited within oral cavity.
The distribution and evolution of CV-A16 had significant epidemiological value to HFMD and HA, and certain value of reference in clinical practice. VP1 region is usually used for serotyping, in which the viral nucleic acid was reversely transcripted, amplified, sequenced, and analyzed by NCBI BLAST. To analyze genotype, subgenotype, and clade determination, we should use phylogenetic tree analysis. There are usually four genotypes, A, B, D, and E of CV-A16 based on VP1 gene while genotype B is divided into four subgenotypes, that is, B1–B4, and subgenotype B1 is again divided into 5 clades of B1a, B1b, B1c, B1d, and B1e. Presently B1a and B1b strain are dominant clades of CV-A16 in the world.5 There were few reports in China on B1c clade cases, most of them were sporadic cases.6-8 In 2022, multiple cases related to CV-A16 of B1c clade were detected in Beijing for the first time. The aim of the study is to provide basic data and reference for the prevention and control of the B1c strains.
2 METHODS
2.1 Surveillance system
Since 2008, a comprehensive surveillance network has been in place in Beijing, comprising 29 hospitals and 16 district Centers for Disease Control (CDCs). Clinical cases of HFMD were diagnosed in accordance with the Hand, Foot and Mouth Disease Diagnosis and Treatment Guide (version 2018),1 with oral vesicular exanthema/ulcers on the tongue or the buccal mucosa, and vesicular rashes over the palms, soles, buttocks or the trunk. HA clinical cases were diagnosed according to the Expert Consensus on Diagnosis and Treatment of Herpangina (version 2019).9
This program included the first five outpatients diagnosed with HFMD and HA each month across the participating 29 hospitals. Samples, including throat swabs and/or stool/rectal swabs, were collected from HFMD and HA cases by trained technicians. Total RNA was extracted directly from 200 μL of each sample by QIAamp Viral RNA mini kit (QIAGEN, cat. no. 52906). Detection of enteroviruses was performed using Real-time RT-PCR (Nucleic Acid Detection Kit with Triple Channels for EV71/CA16/EV, Guangzhou Daan Science and Technology Co. Ltd., cat. no. DS0932) for both detection of enterovirus with universal primers and probe and to determine serotypes of EV71 and CV-A16. For each month, each district CDC selected and sent 2–4 CV-A16 positive samples to Beijing CDC for full-length sequencing of VP1 gene.
2.2 Genotyping with VP1 gene and sequence analysis
The extract RNA of the subjects were used for amplification of VP1 gene by semi-nested RT-PCR. The primers10, 11 were listed in Table 1 and synthesized by Takara Biotechnology (Dalian) Co. Ltd. The primer pair of HEVAS1495 and HEVAR2C and SuperScript™ III one-step RT-PCR kit (Invitrogen, Art. No. 12574-026) were used in the reverse transcription and first round of amplification. The conditions were 45 min at 42°C, and 3 min at 94°C, followed by 40 cycles of 30 s at 94°C, 45 s at 45°C and 2.5 min at 68.5°C, and a final extension of 5 min at 68.5°C. The primer pair of HEVAS1495 and HEVAR2807 were used in a second PCR amplification. The conditions were 3 min at 94°C, and followed by 30 cycles of 30 s at 94°C, 45 s at 45°C, and 1.5 min at 72°C, and a final extension of 5 min at 68.5°C. The PCR products were analyzed on a QIAxcel capillary electrophoresis instrument (QIAGEN) and the PCR products were sequenced by Shanghai Invitrogen Co. Ltd.
| Primer name | Sequence(5'−3') |
|---|---|
| HEVAS1495 | CAGTCTTCATCAGTTACCCTGGTIATICCUGGATIAGYAACAC |
| HEVAR2C | CGGTGYTTGCTCITGAACTGCATG |
| HEVAR2807 | CAGTTACACCGGGCAATCGTGTCRCAICCYTGIGCIGTRG |
The paired sequences were aligned and assembled into one consensus sequence. NCBI BLAST was used to determine serotypes. MEGA 10.0 software was used for multiple alignment of the VP1 sequences and 216 representative sequences of CV-A16 from NCBI. Selection of representative sets of CV-A16 VP1 sequences was described by Li et al.12, 13 In brief, the 216 representative VP1 sequences encompassed isolates from 25 different countries and spanned a period from 1951 to 2020 and were selected from distinct branches of the neighbor-joining (NJ) phylogenetic tree of all VP1 sequences of CV-A16 in GenBank. The selection process was designed to ensure that the data set was as representative as possible, taking into account the collection country and date characteristics of the isolates.
The phylogenetic tree of NJ method was construction for determination of genotype, subgenotype, and clade. Pairwise identity score (PIS) was used to describe the similarity within or between genotypes/subgenotypes/clades.13 PIS as 1.00 represented 100% similarity. The distribution of the similarity was visualized by Graphpad Prism 8.
2.3 Genomic sequencing of B1c strains
Genomic sequences of B1c stains were obtained by next-generation sequencing (NGS) using genome amplification products. The primers to amplify long segments were designed according to 27 reference genomes of B1c stains downloaded from NCBI database and the sequences of the primers are listed in Table 2. CLC Genomics Workbench version 10.1.1 software (QIAGEN) was used for sequencing data analyses, including quality control, trimming of low-quality data, genome assembly, and alignment of reads and contigs with local nonredundant nucleic acid database using BLASTn.
| Primer set | Primer | Nucleotide sequence | Locationa |
|---|---|---|---|
| 1 | F1: | TTAAAACAGCCTGGGGGTTGTTCCC | 1–27 |
| R1475: | TTAATTGACTCAAAGGGATCCCTGC | 1475–1455 | |
| 2 | F1210: | TGACAGAAGTAGGTGTTTTTGGTC | 1210–1233 |
| R2704: | CCAGCACGGCTAAAGAAATTCCC | 2704–2682 | |
| 3 | F2319: | TATGTGGTACCCATTGGTGCTCC | 2319–2341 |
| R3867: | TCAATGCTTCCACTTCCCTGGACAC | 3867–3843 | |
| 4 | F3457: | TCTCTTCCACCACCGCCCAGGGGTG | 3457–3481 |
| R4947: | TGGCTTTGCCACACACGAGTGGGCT | 4947–4923 | |
| 5 | F4542: | TACTCACTCCCTCCAGATCCAGACC | 4542–4566 |
| R5936: | TTCACTGGCAAAGTAACCCCTCTT | 5936–5913 | |
| 6 | F5712: | CACATGCCCTCAATGTTCGTTCC | 5712–5734 |
| R7409: | TACTTCTGGTTATAACAAATTTACC | 7409–7385 |
- a Location according to CV-A16 strain of B1c clade (MH780757.1).
The whole genome sequences obtained in this study and 400 whole genome sequences downloaded from NCBI were constructed by NJ method with 1000 bootstrap replicates.
2.4 Amino/nucleic acid frequency differences in clades of B1a–B1e
The relative frequency of amino/nucleic acid at each position of amino/nucleic acid sequence are visualized by sequence logo (WebLogo v3;http://weblogo.threeplusone.com/).14 The height of the symbol represents the probability of amino acid or nucleic acid at the position. Visualization by logo is used to compare the difference between the complete VP1 sequences of the studied stains and stains of five clades, including B1a–B1e.
2.5 Analysis of epidemiological relationship among B1c cases
The basic information, disease onset time, hospital visit, and testing results of the study subjects were collected. A diagram of geographic location of roads, communities, schools, kindergartens related to B1c cases as well as the scale were copied from online map. The information on the cases as well as the area were added into the diagram for the analysis of the relationship among cases.
3 RESULTS
3.1 Genotyping results of the cases
In 2022, a total of 893 clinical cases, comprising 400 HFMD cases and 493 HA cases, were assembled from the sentinel hospitals and collected by the district CDCs. Subsequent real-time RT-PCR testing identified 368 samples that tested positive for enteroviruses. Among these, 180 enterovirus-positive cases—specifically, 131 HFMD cases and 49 HA cases—were forwarded to the Beijing CDC for further analysis. Thirty-four samples (18.89%) were identified as CV-A16 infections, EV-A71 was present in a solitary case (0.56%), CV-A6 was the predominant strain, comprising 114 of the isolates (63.33%). Additionally, 31 strains (17.22%) were attributed to other enteroviruses. Full-length VP1 gene sequences were amplified and sequenced successfully in 22 cases. The serotypes were confirmed as CV-A16 by NCBI BLAST. A phylogenetic analysis was conducted using the 22 sequences, which revealed that the CV-A16 strains isolated from the Beijing region in 2022 were localized to the evolutionary branch characteristic of the B1 genetic subtype. Specifically, the B1a, B1b, and B1c clades were represented by 14, 3, and 5 cases, respectively. Consequently, the B1a subtype was the most prevalent among the CV-A16 strains in Beijing for 2022, with the B1b and B1c subtypes also identified.
3.2 Phylogenetic analysis of B1c stains
The B1c clade was categorized into three distinct branches—Malaysia, Russia/France, and India—based on analyses of VP1 sequences (Figure 1A) and complete genomic sequences (Figure 1B). The Malaysia branch exclusively contained strains from Malaysia, predominantly isolated between 2005 and 2018. The Russia/France branch comprised mainly strains from Russia (2009–2011) and France (2010–2011), including a strain from Kazakhstan in 2011.
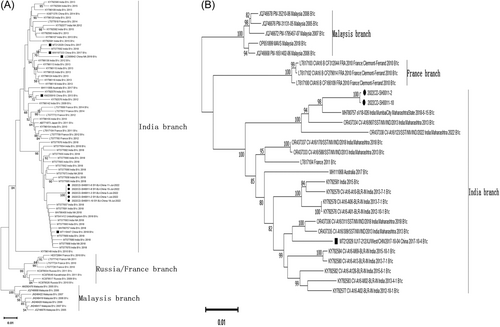
The India branch demonstrated a broader temporal distribution, with strains isolated annually from India between 2009 and 2022. It also included strains from Japan in 2011, France from 2011 to 2014, Australia in 2017, China from 2013 to 2019, the United Kingdom in 2018, and the B1c strains discovered in the Beijing region during this study. These latter strains are represented by five complete VP1 gene sequences and two comprehensive genomic sequences.
The five complete VP1 gene nucleotide sequences from the B1c clade strains identified in Beijing were subjected to NCBI BLAST analysis, which demonstrated the highest sequence identity with the Indian strain oV18-026 isolated in 2018 (GenBank accession number MH780757.1), with a similarity of 98.09%.
Of these, two viral strains exhibited full-genome sequences that were genetically most similar to the 2018 Indian oV18-026 strain, and the BLAST analysis indicated the closest genetic resemblance to this strain, with similarities of 98.09% and 98.11%, respectively.
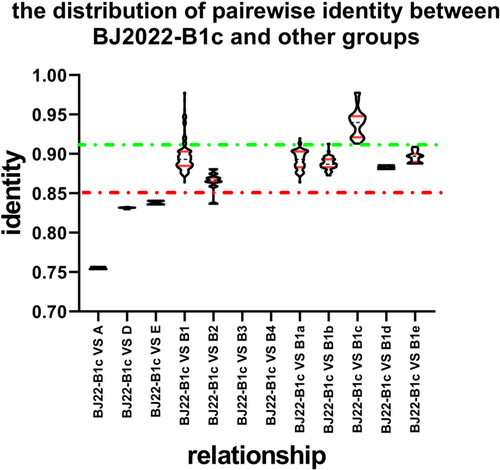
3.3 Similarity analysis
The five procured full-length VP1 gene sequences, along with 216 representative reference sequences, were employed for a comparative similarity analysis across genotypes/subgenotypes. As depicted in Figure 2, PIS of nucleotide sequences revealed that the median PIS values for the five B1c strains in Beijing with genotypes A, D, and E, were 0.756, 0.832, and 0.839, respectively. Within genotype B, the median PIS values for these five B1c strains with strains classified into subgenotypes B1 through B4 were 0.893, 0.867, 0.878, and 0.879, respectively. Among the clades classified within the B1 sub-genotype, the PIS values for the strains associated with B1a to B1e were 0.893, 0.887, 0.940, 0.883, and 0.897, respectively.
3.4 Amino/nucleic acid frequency analysis
In addition to similarity and evolutionary analyses, we also examined the frequencies of amino and nucleic acids in the sequences of the isolated B1c strains, the representative strains of B1c and other clades within the B1 subgenogroup. Among the sequences of the five B1c strains from Beijing, there were 27 specific nucleotide mutations characteristic of B1c, such as 7T, 138T, 366G, 471T, 474G, 543T, 591G, 621A, 703G, 711G, 718G, 744A, and 822A (as illustrated in Figure 3). Additionally, there were three unique nucleotide mutations in the Beijing-isolated strains: 74C, 465C, and 843T.
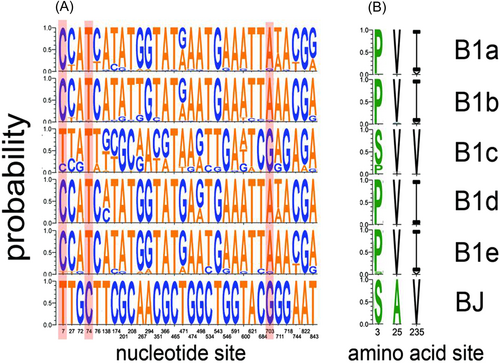
Regarding the aforementioned specific nucleotide mutations, only three amino acids at positions 7T, 74C, and 703G were altered in the encoded sequence (as indicated by the shaded area in Figure 3A), corresponding to amino acids 3S, 25A, and 235V, respectively (Figure 3B). While the amino acid 25A represents a unique mutation identified in this study, the other two correspond to common mutations found within the B1c clade.
3.5 Basic spatiotemporal analysis of the B1c cases
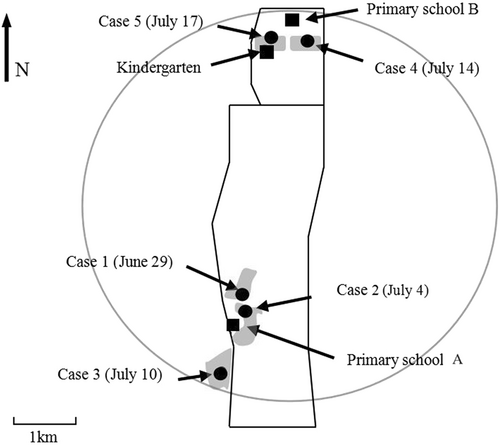
Shunyi District reported five cases of the B1c clade CV-A16 virus strains. Upon investigation, the five cases exhibited a certain degree of spatiotemporal clustering.
A summary of the case details can be found in Table 3. The onset dates for the cases spanned from June 29 to July 17. Commencing with case 1, the intervals between the subsequent onset dates were 5 days, 6 days, 4 days, and 4 days, respectively. All patients were male. Regarding age demographics, four of the patients (cases 1, 2, 3, and 5) fell within the primary school age range of 8–10 years, whereas one patient (case 4) was a preschooler at 4 years of age. From a clinical perspective, the symptoms experienced by the cases were classified as mild, characterized by the presence of herpetic lesions and the absence of fever.
| Case | Age | Gender | Distance to case 1 (km) | Community | Disease onset | Clinical feature | Fever | Rash |
|---|---|---|---|---|---|---|---|---|
| 1 | 10.04 | Male | 0 | WQ | 29/Jun/2022 | Mild | No | Yes |
| 2 | 10.22 | Male | 0.2 | WQ | 4/Jul/2022 | Mild | No | Yes |
| 3 | 10.06 | Male | 1.5 | WQ | 10/Jul/2022 | Mild | No | Yes |
| 4 | 4.49 | Male | 5 | MP | 14/Jul/2022 | Mild | No | Yes |
| 5 | 8.11 | Male | 5 | MP | 17/Jul/2022 | Mild | No | Yes |
- Abbreviations: MP, Mapo community; WQ, Wangquan community.
As illustrated in Figure 4, geographically, all patients resided within a region that had a diameter of 7 km. Additionally, the cases were clustered into two distinct neighborhood groups. Cases 1, 2, and 3 were located in neighboring areas in the southern part of the region, approximately 1.5 km apart. Cases 4 and 5 were situated in close proximity in the northern part, about 1 km apart. The distance separating the two neighborhood clusters was 5 km.
4 DISCUSSION
CV-A16 is the common pathogen causing HFMD in China, and the main epidemic trend of co-circulation of B1a and B1b clades. From 2013 to 2018 B1b clade was the dominant type. From 2019, the incidence of B1a clade increased.5, 15 There was also co-circulation in Beijing but the dominant type was B1a clade.12 In the previous reports, B1c clade was detected in France and Japan around 2010.16 The epidemics and wide spreading of the virus occurred in India during 2009–2018.17, 18 In 2017 and 2018, the B1c clade strains were identified in Australia (MH111068) and United Kingdom (MT641412). The reports of B1c cases in China were rather sporadic, including the case in Shanghai in 2014, the case in Guangzhou in 2018 and the case in Yunnan in 2019.6-8 This study presents the first report from China on multiple related cases of HFMD caused by the CV-A16 B1c clade.
Beijing has had an extensive period for the detection of enteroviruses, initiating surveillance for HFMD in 2008,3 and gradually incorporating outbreaks of fever and HA cases into the surveillance system. Past surveillance has detected serotype replacements,19 providing a reference for the development of the EV-A71 vaccine and for prevention and control measures in Beijing. In recent years, the Beijing area has exhibited a co-circulation of the B1a and B1b genotypes; however, the B1a genotype has been identified as the predominant strain in this region.12 Previous literature has reported the identification of one case of B1c in a hospital in Beijing in 201620; however, it was not confirmed whether it was a local case in Beijing. Therefore, B1c remains quite rare in Beijing, as it is in other parts of the country. In 2022, for the first time in Beijing, consecutive cases of B1c-positive HFMD occurred within two communities in Shunyi District.
The genetic sequences acquired in this study indicate a close phylogenetic proximity and substantial alignment with Indian strains according to both evolutionary distance analysis and BLAST results, albeit with certain discrepancies. We hypothesize that mutations may have arisen throughout the transmission process. However, in the absence of concrete evidence for such variations, heightened scrutiny and surveillance of the B1c clade are warranted.
Analysis of amino acid variation sites revealed that there were mutations at three amino acid positions within the VP1 coding region. At the 3rd amino acid position, all five strains exhibited a P-S mutation; at the 235th position, all B1c strains showed an I-V mutation. These mutations have also been reported in recent studies on CV-A16 in China,21, 22 suggesting that these two positions may have evolved into more stable genetic traits that are being passed on. Additionally, a unique local amino acid change occurred at the 25th position (V-A). Current research on amino acid variation sites has primarily focused on the B1a and B1b clusters, with fewer reports on B1c. Whether these amino acid variations directly affect viral antigenicity requires further investigation.
This study constitutes a retrospective investigation, with a limitation being that the five cases were derived from surveillance at sentinel hospitals, lacking more detailed epidemiological information. Based solely on the available spatiotemporal data, it is not sufficient to conclusively demonstrate that they originated from the same outbreak.
In summary, the B1 subgenotype of CV-A16 is one of the main etiological agents of HFMD and HA in the Beijing area. For the first time in China, multiple cases of CV-A16 virus of B1c clade within a short period occurred in Beijing's Shunyi District.
Therefore, it is necessary to enhance the monitoring of the B1c clade CV-A16 in HFMD and HA in this region. This includes the associated clinical symptoms following infection, as well as the evolution and variation at the genetic level. Such surveillance can provide a basis for future prevention and control measures.
AUTHOR CONTRIBUTIONS
Zhichao Liang, Renqing Li, and Changying Lin carried out the experimental design, participated in the experiment, and drafted the manuscript. Da Huo, Xiangfeng Dou, Shuaibing Dong, Dan Li, Zhenyong Ren, and Yulan Sun participated in the experimental design, performed the data analysis, and reviewed the manuscript. Yang Yang, Shujuan Cui, Zhaomin Feng and participated in the experiment and reviewed the manuscript. Dan Wu has uploaded the full genomic nucleotide sequence data to the public database NCBI. Lei Jia and Zhiyong Gao contributed to the experimental design and coordination with District Centers for Disease Control and Prevention. Renqing Li provided a final review of this manuscript. All authors read and approved the final manuscript.
ACKNOWLEDGMENTS
We thank all participants who provided samples and clinical data. We also thank the staff of 16 district CDCs in Beijing and clinical staff for collecting epidemiological information and samples during the execution of this study. None of the funders had any influence over the study design, analysis, or decision to submit for publication.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ETHICS STATEMENT
This study adhered to the Helsinki Declaration and was approved by the Human Research Ethics Committee of Beijing Center for Disease Control and Prevention (Beijing CDC). Informed consent was obtained from either the patient or the patient's guardian for sample collection.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/nuccore, reference numbers PP730024 and PP730025. The complete genomic sequences of two CV-A16 strains obtained in this study were submitted to NCBI (accession numbers: PP730024, PP730025).




