Characterization of CRESS-DNA viruses in human vaginal secretions: An exploratory metagenomic investigation
Endrya do Socorro Foro Ramos, Tania Regina Tozetto-Mendoza, and Pietro Bortoletto contributed equally to this work.
Elcio Leal, Antonio Charlys da Costa, Steven S. Witkin, Maria Cassia Mendes-Correa jointly supervised this work.
Abstract
The Phylum Cressdnaviricota consists of a large number of circular Rep-encoding single-stranded (CRESS)-DNA viruses. Recently, metagenomic analyzes revealed their ubiquitous distribution in a diverse range of eukaryotes. Data relating to CRESS-DNA viruses in humans remains scarce. Our study investigated the presence and genetic diversity of CRESS-DNA viruses in human vaginal secretions. Vaginal swabs were collected from 28 women between 29 and 43 years old attending a fertility clinic in New York City. An exploratory metagenomic analysis was performed and detection of CRESS-DNA viruses was confirmed through analysis of near full-length sequences of the viral isolates. A phylogenetic tree was based on the REP open reading frame sequences of the CRESS-DNA virus genome. Eleven nearly complete CRESS-DNA viral genomes were identified in 16 (57.1%) women. There were no associations between the presence of these viruses and any demographic or clinical parameters. Phylogenetic analysis indicated that one of the sequences belonged to the genus Gemycircularvirus within the Genomoviridae family, while ten sequences represented previously unclassified species of CRESS-DNA viruses.
Novel species of CRESS-DNA viruses are present in the vaginal tract of adult women. Although they be transient commensal agents, the potential clinical implications for their presence at this site cannot be dismissed.
1 INTRODUCTION
The phylum Cressdnaviricota consists of a large number of viruses with small, circular, single-stranded DNA genomes encoding replication-associated proteins with an HUH endonuclease domain and a SH3 superfamily helicase domain. It encompasses at least 12 established families (Bacilladnaviridae, Circoviridae, Vilyaviridae, Smacoviridae, Amesuviridae, Metaxyviridae, Nanoviridae, Redondoviridae, Naryaviridae, Nenyaviridae, Geminiviridae and Genomoviridae). These families as well as several other unclassified circular Rep-encoding single-stranded (CRESS)-DNA viruses have been described based on phylogenetic analysis of Rep protein sequences.1-3 These viruses are ubiquitous and have been identified in a wide variety of eukaryotic species as well as in environmental samples.1-5 Two decades ago, the application of phi29 DNA polymerase and random hexamers (rolling circle amplification; RCA) for whole genome amplification of circular DNA templates heralded a turning point in the study of CRESS DNA viruses.6, 7 Nowadays, analysis of metagenomic data has increased our understanding of the complex genetic diversity of CRESS-DNA viruses, culminating in the creation of the Cressdnaviricota phylum in 2019 by the International Committee on Taxonomy of Viruses.8 Taxonomic changes in these families have continuously been outlined.9 Many CRESS-DNA viruses have been implicated as causative agents of economically important animal disease.2 Their presence has recently been observed in human samples such as pericardial fluid, feces, cerebrospinal fluid, blood and respiratory secretions. However, causal evidence of the relationship between their presence and human abnormalities or disease remains to be elucidated.10-14 In the present study we evaluated by metagenomic analysis the presence and diversity of CRESS-DNA viruses in vaginal secretions from reproductive age women attending a fertility clinic.
2 MATERIALS AND METHODS
2.1 Setting and study group subjects
Women seen for an initial consultation at The Center for Reproductive Medicine and Infertility at Weill Cornell Medicine in New York City were informed of our intention to investigate the presence of viruses in vaginal secretions. Twenty-eight women agreed to participate and provided an initial sample. A second sample was collected from 21 of these women 12–21 days later. Following obtaining written informed consent, samples were collected from the vaginal walls with a sterile cotton swab during a routine speculum-based vaginal examination. The swab was vigorously shaken into a tube containing 1 mL of sterile phosphate-buffered saline, the swab was removed and the sample was stored at −80°C before being shipped on dry ice to the virology laboratory (LIM-52) at the Faculdade de Medicina da Universidade de São Paulo (FMUSP) in São Paulo, Brazil. All samples arrived intact and still frozen. Clinical and demographic data were subsequently obtained by chart review. The study was approved by the Institutional Review Board at Weill Cornell Medicine, under protocol number 20-04021885 in October 2022.
2.2 Methodology
The samples were thawed, transferred into a 2 mL impact-resistant tube containing lysing matrix D (MP Biomedicals) and 1 mL sterile Hank's Balanced Salt Solution and homogenized in a FastPrep-24 5G (MP Biomedicals). The homogenized sample was centrifuged at 8000g for 10 min and approximately 500 μL of the supernatant was then percolated through a 0.45 μm filter (Merck Millipore) to remove eukaryotic and bacterial cell-sized particles. Use of this filter size allowed the performance of a thorough exploratory metagenomic approach to facilitate the capture of a more complete spectrum of the virus community that may have been present.15-19 Approximately 100 μL, roughly equivalent to one fourth of the volume of the tube, of cold PEG-it Virus Precipitation Solution (System Biosciences) was added to the filtrate and the contents of the tube were gently mixed and then incubated at 4°C for 24 h. After the incubation period, the mixture was centrifuged at 8000g for 30 min at 4°C. Following centrifugation, the supernatant (~350 μL) was discarded. The pellet, rich in viral particles, was treated with a combination of nuclease enzymes (TURBO DNase and RNase Cocktail Enzyme Mix (Thermo Fischer Scientific) Baseline-ZERO DNase (Epicentre) Benzonase, (Merck Millipore) and RQ1 RNase-Free DNase (Promega) to digest unprotected nucleic acids. The resulting mixture was subsequently incubated at 37°C for 2 h. Immediately after treatment, total nucleic acids were extracted using the automated Maxwell RSC Viral Total Nucleic Acid Purification Kit (Promega). A first round of complementary DNA (cDNA) synthesis was performed with Superscript IV Reverse Transcriptase (Thermo Fisher Scientific). A second strand cDNA synthesis was performed using a DNA Polymerase I Large (Klenow) Fragment (Promega), purified with ProNex Size-Selective Purification System (Promega) and submitted to fluorometric quantification with the QuantiFluor ONE dsDNA System (Promega).
Viral DNA was enriched for circular DNA molecules using RCA with the Illustra TempliPhi kit (Cytiva), purified with ProNex Size-Selective Purification System (Promega) and submitted to fluorometric quantification with QuantiFluor ONE dsDNA System (Promega). The samples were assayed in duplicate for total nucleic acid and for aliquots enriched for viruses with circular genomes.
Subsequently, 2 ng of DNA was submitted to the Nextera XT Sample Preparation Kit (Illumina) and used to construct a DNA library, which was identified using dual barcodes. For size range, Pippin Prep (Sage Science, Inc.) was used to select a 500 bp insert (range: 400–600 bp). The library was deep-sequenced using a NovaSeq. 6000 Sequencer (Illumina) with 2 × 250 bp ends.
2.3 Alignments and annotation
The resulting contigs were subjected to a modified protein BLAST search using Ugene software20 to identify novel members of CRESS-DNA viruses. Based on the best results (best hits) from BLASTx search, genomes of CRESS-DNA viruses and other related viruses were chosen for further analysis. Full or nearly full genomes were aligned using MAFFT software.21 Genome annotation was performed using Gatu software22 with the CRESS-DNA virus (CRESSV) sequence AUM61914 as reference. The translated sequences were used to determine viral motifs in the online server Motif Finder (https://www.genome.jp/tools/motif/).
2.4 Genetic identity
Genetic distances and their standard error were calculated using the maximum composite likelihood model plus gamma correction and bootstrap with 100 replicates. Distances were calculated using MEGA software (Version X).23 To estimate the similarity of sequences we used a pair-wise method implemented in the program SDT.24 The similarity alignments of every unique pair of sequences were estimated using algorithms implemented in MUSCLE.25 After computation of the identity score for each pair of sequences the program then used the NEIGHBOR component of PHYLIP to compute a tree.26 The rooted neighbor-joining phylogenetic tree ordered all sequences according to their likely degree of evolutionary relatedness. Results are presented in a frequency distribution of pairwise-identities in a graphical interface.
2.5 Phylogenetic analysis
Phylogenetic trees based on the ORF 2 (REP) of CRESSV genomes were constructed using the maximum likelihood approach and branching support was estimated using an ultrafast bootstrap test with 1200 replications using the IQ-TREE tool.27 The tree was visualized and edited using Figtree version 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree). The viral genomes described in detail here were deposited in GenBank.
3 RESULTS
Previously unclassified and potentially novel species of CRESS-DNA viruses were detected in the vaginal tract of adult women. A total of 11 near full length sequences of CRESSV were detected in vaginal samples from 16 (57.1%) study participants. In the 21 women from whom vaginal samples were collected at two different time periods, nine (42.9%) of the first samples and five (23.8%) of the second samples were CRESSV-positive. Four (57.1%) of the seven women with only one collected vaginal sample were also CRESSV-positive. A total of seven vaginal samples were positive for V05, five each were positive for V09 or V10, two were positive for V07, and one sample each was positive for V12, V13, V14, V15, V16, or V17. Another vaginal sample was positive only for Genomovirus. In only two cases were both samples from the same woman positive for CRESS-DNA, but different viruses were detected in each sample. These results are shown in Tables S1 and S2. There were no differences between women who were positive (n = 16) or negative (n = 12) for CRESSV in terms of age, body mass index, race, gravidity, parity, fertility status, or cause of infertility (Table 1).
| CRESS positive | CRESS negative | |
|---|---|---|
| Parameter | n = 16 | n = 12 |
| Mean age (SD) | 36.9 (5.1) | 36.6 (4.2) |
| Mean BMI (SD) | 25.4 (5.3) | 26.1 (7.8) |
| Median gravidity (IQR) | 1.0 (0,2) | 0 (0,2.5) |
| Median parity (IQR) | 0 (0,0) | 0 (0,0) |
| Race | ||
| White | 10 (62.5%) | 7 (58.3%) |
| Non-White | 6 (37.5%) | 5 (41.7%) |
| Fertility problem | ||
| None | 2 (12.5%) | 3 (25.0%) |
| Poor ovulation | 7 (43.8%) | 5 (41.7%) |
| Male factor | 2 (12.5%) | 2 (16.7%) |
| Idiopathic | 3 (18.8%) | 1 (8.3%) |
| Tubal | 0 | 1 (8.3%) |
| Endometriosis | 1 (6.3%) | 0 |
- Abbreviations: BMI, body mass index (kg/m2); CRESS, circular Rep-encoding single-stranded; IQR, interquartile range; SD, standard deviation.
The amino acid identities of the different isolates of CRESS-DNA viruses are shown in Table 2. A comparative analysis using the BLASTX tool for homology search revealed that the sequences shared 40%–68% identity with reference sequences present in the database. Amino acid identity is related to the cognate REP genomic region of CRESSV reference sequences deposited in Genbank and is based on BLASTX search.
| CRESSVa | Size (bp) | Reference Genbank | Coverage | E-values | Identity* |
|---|---|---|---|---|---|
| V05 | 1948 | AUM61914 | 53% | 1,00E-67 | 49.13% |
| V17 | 2257 | AUM61914 | 56% | 3,00E-67 | 49.20% |
| V09 | 2100 | AUM61914 | 40% | 2,00E-67 | 49.13% |
| V12 | 2032 | AUM61914 | 40% | 2,00E-67 | 49.13% |
| V10 | 1869 | AUM61914 | 44% | 2,00E-67 | 49.13% |
| V13 | 1839 | AUM61914 | 40% | 1,00E-61 | 49.03% |
| V14 | 1543 | AUM61914 | 34% | 9,00E-48 | 44.43% |
| V07 | 1753 | AUM61914 | 49% | 8,00E-61 | 40.34% |
| V15 | 1542 | AXH76771 | 34% | 9,00E-48 | 44.43% |
| V16 | 1409 | AXH76771 | 39% | 8,00E-50 | 44.67% |
| Gemycircularvirus | |||||
| Gen_V01 | 2906 | YP_010798304 | 31% | 9,00E-114 | 68.06% |
- Abbreviation: CRESS, circular Rep-encoding single-stranded.
- a Genbank accession numbers of CRESS-DNA virus (CRESSV) sequences of this study: Gen_V01 (PP750555); V05 (PP750567); V07 (PP750569); V09 (PP750568); V10 (PP750563); V12 (PP750566); V13 (PP750565); V14 (PP750558); V15 (PP750559); V16 (PP750560); V17 (PP750556). *consulted identidy in genBank.
- * consulted identidy in genBank.
We employed the ORFfinder program (https://www.ncbi.nlm.nih.gov/orffinder/), to conduct a predictive analysis of open reading frames (ORF) within our sequences. The outcomes are summarized in Table 3, detailing the positions of ORF1 (CAP) and ORF2 within the genome of CRESS viruses. It is important to note that the complete CAP comprises 798 nucleotides, while REP spans 843 nucleotides. Notably, some sequences exhibit incomplete ORFs due to the presence of stop codons.
| Position in the genome (size nucleotides) [frame sense] | ||
|---|---|---|
| CRESSVa | ORF 1 (CAP) | ORF 2 (REP) |
| V05 | 454-913 (450) [−] | 1917-1075 (843) [+] |
| V17 | 442-1239 (798) [+] | 2243-1401 (843) [−] |
| V09 | 273-1070 (798) [+] | 2074-1232 (843) [−] |
| V12 | 8-805 (798) [+] | 1809-967 (843) [−] |
| V10 | 19-816 (798) [+] | 1820-978 (843) [−] |
| V13 | 34-831 (798) [+] | 1739-993 (747) [−] |
| V14 | 207-1004 (798) [+] | 1636-1166 (471) [−] |
| V07 | 14-682 (669) [+] | 1686-844 (843) [−] |
| V15 | 40-837 (798) [+] | 1469-999 (471) [−] |
| V16 | 17-685 (669) [+] | 1317-846 (470) [−] |
| Gen_V1 | 1659-742 (918) [−] | 1817-2393 (549) [−] |
- Abbreviation: CRESS, circular Rep-encoding single-stranded.
- a Genbank accession numbers of CRESS-DNA vírus (CRESSV) sequencies of this study: Gen_V01 (PP750555); V05 (PP750567); V07 (PP750569); V09 (PP750568); V10 (PP750563); V12 (PP750566); V13 (PP750565); V14 (PP750558); V15 (PP750559); V16 (PP750560); V17 (PP750556).
To aid in visualization, Figure 1A,B depicts genome maps of CRESSV sequences V09 and V01, respectively. These figures also highlight the amino acid sequences of CAP, emphasizing a substantial abundance of positively charged amino acids, namely lysine (K) and arginine (R), within the amino terminal region. These positively charged amino acids play a crucial role in the virus's DNA packaging process.28
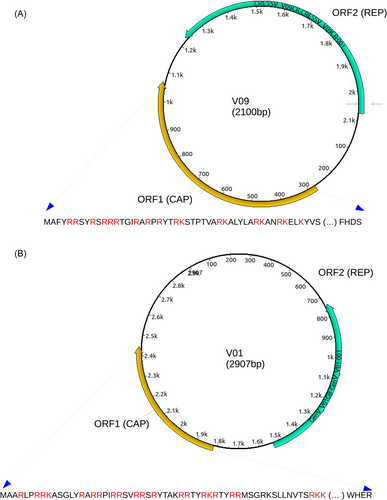
It is important to highlight that we observed variations of the Kozak motif (A/G)CCaugG in the start codon regions of CAP and REP within the genomes of CRESSV and V01. Specifically, the CRESS-DNA genome featured the motif GCAaugG for CAP and TCCaugA for REP, while GenV01 exhibited the motif ACAaugG for CAP and CCAaugC for REP (Figure 1). The Kozak consensus sequence is a pivotal nucleic acid motif prevalent in the majority of eukaryotic messenger RNA transcripts, playing a central role in initiating protein translation within eukaryotic cells.
To determine the evolutionary history of the DNA sequences relative to other CRESSV, we used the REP protein from the sequence of this study and other references classified in the phylum Cressdnaviricota, class: Arfiviricetese (Amesuviridae, Bacilladnaviridae, Circoviridae, Metaxyviridae, Nanoviridae, Naryaviridae, Nenyaviridae, Redondoviridae, Smacoviridae, Vilyaviridae) and Repensiviridae (Geminiviridae, Genomoviridae).29
In the tree, the sequences grouped robustly (100% Bootstrap) into two clades (Figure 2). In clade I, the CRESSV sequences (i.e., V05, V17, V09, V12, V10, V13, V14, V07, V15, V16) exhibited greater relationship to unclassified circular DNA virus sequences (AUM61914) identified in wastewater in 2015 in the United States (by Pearson, V.M. and Rokyta, D.R from Department of Biological Science, Florida State University, Tallahassee, USA).
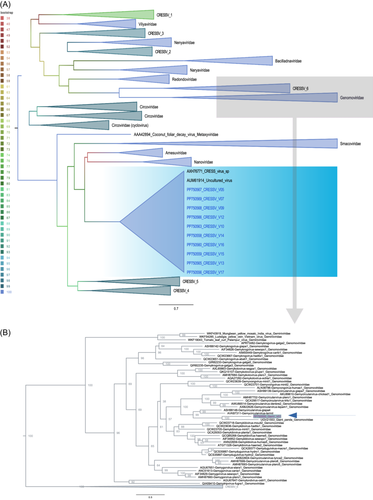
To gain deeper insights into phylogenetic relationships between the sequences, we conducted pairwise comparisons, focusing on the REP region at the amino acid level. The expected results revealed a high degree of identity, ranging from 97% to 100% when compared to each other. However, when compared to reference sequences AUM61914 (by Pearson, V.M. and Rokyta, D.R. from Department of Biological Science, Florida State University, Tallahassee, USA) and AXH76771 (by Tisza, M., Buck, C., Pastrana, D., Welch, N. and Peretti, A. Bethesda, USA), the identity dropped to 44%–50% and 37%–43%, respectively.
In the context of phylogenetic classification, a notable correlation, ranging from 31% to 39%, was evident upon comparing sequences with those derived from the Nanoviridae family. Conversely, within the CP (coat protein) region, a remarkable consistency was observed among the sequences, displaying a complete 100% identity amongst themselves. This contrasted sharply with their 27% identity alignment with the AUM61913 sequence (by Pearson, V.M. and Rokyta, D.R from Department of Biological Science, Florida State University, Tallahassee, USA) and a 22% identity correspondence with AXH76770 (authored by Tisza, M., Buck, C., Pastrana, D., Welch, N., and Peretti, A. from Bethesda, USA) and sequences affiliated with the Nanoviridae family.
These findings emphasize the remarkable breadth of genetic variation present among CRESS-DNA viruses. The sequences of CRESS-DNA analyzed in this study exhibited a lack of alignment with any established taxonomic family within the Cressdnaviricota phylum, suggesting the presence of novel evolutionary lineages.
Clade II includes V01, which showed the greatest relationship with the unclassified Giant Panda sequence Genomoviridae UGV21553 (by Lu, X., Yang, X. S., and Zhang, W. from School of Medicine, Jiangsu University, Xiangshan, China) detected in an anal swab and other members classified in the family Genomoviridae genus Gemycircularvirus. Paired comparisons in REP at the amino acid level resulted in 66% identity with the UGV21553 sequence and 44%–52% with the other Gemycircularvirus sequences. Considering the species demarcation criteria established for Genomoviridae >78% in the complete genome.30 V01 is probably a new species within the genus. It shares 65% identity with Giant Panda Genomoviridae (MZ956591) and 62%–66% with the species Gemycircularvirus-giapa4 (MF327561) and Gemycircularvirus-giapa3 (MF327568), respectively.9, 31
In relation to the detection of RNA viruses, when present their genomes were represented by only a low number of reads in our study, and without our availability for independent confirmation.
4 DISCUSSION
The unique sequences of CRESS-DNA viruses present in the vagina of reproductive age women in The United States are described for the first time. The present study identified 11 near full-length sequences of these CRESS-DNA isolates that represent unclassified species. This favored their genetic characterization as novel CRESS-DNA viruses belonging to the Cressdnaviricota phylum. There has been a paucity of whole-genome sequences for eukaryote-infecting CRESS-DNA viruses isolated from the female genital tract. Currently, both the Metagenomic tools and the rolling-circle replication methods for sequence amplification have favored the identification and annotation of a repertory of novel CRESS-DNA viruses and the discovery of their range of eukaryotic hosts.6, 7, 32 Our findings underscore the substantial diversity within CRESS-DNA viruses. Notably, the CRESSV-DNA sequences that we describe do not align with any established family within Cressdnaviricota, supporting that they might constitute a novel group of CRESS-DNA viruses.
According to the ICTV, CRESS-DNA viruses have genomes between 1.7 and 6.0 kb. Genomes from the 11 CRESS-DNA isolates in the present study ranged from ~1.5k to ~2.5 kb in size. We observed that the presence of CRESS-DNA viruses in the vaginal samples was a transient event, detected in only one of two paired samples from the same woman collected 2–3 weeks apart. Whether this indicates a limitation of the sensitivity of our assay and/or variation in viral expression in human cells or microbial cells co-occupying the vaginal milieu remain to be determined.
Moreover, one woman presented in her vagina sequences of CRESSV which were more closely related to the Genomoviridae family. The presence of this Genomavirus isolate also did not persist within the 11 days between collection of the first and second sample from the same woman. Genomovirus has been identified in a wide spectrum of animal and plants hosts. In phylogenetic reconstruction, the viral sequences of this Genomovirus isolate showed identity with Genomovirus from many other eukaryotic organisms and environmental samples.1-5
Detection of eukaryote-infecting CRESS-DNA viruses in cervical samples from young African adolescent women were reported in the presence of Anelloviridae and Genomoviridae, although most studies of the vaginal virome were limited to report of double-stranded DNA viruses, with an emphasis on HPV genomes.32, 33 In addition, the simultaneous presence of fungi DNA and Genomovirus was found in a subset of cervical swabs from HIV/HPV co-infected women in which both virome and bacteriome composition were analyzed.34 This viral family was recently described in humans, verifying its occasional detection by metagenomic analysis. The well-established Genomoviridae CRESS-DNA family has been reported to infect fungi present in plants.35 This virus family also has the potential to infect fungi present in the cervico-vaginal region of HIV-positive women carrying both Candida albicans and Genomovirus.34 Conversely, two studies reported Genomovirus in cervico-vaginal samples from healthy South African adolescent women.32, 33 In addition, the oral amoeba Entamoeba gingivalis has been identified as a likely host of redondoviruses.36
A limitation of the present study in the analysis of novel CRESS-DNA viruses, including Genomovirus is that we were not able to distinguish if the viruses infected human cells or, alternatively, eukaryotic microorganisms present in the vagina. Furthermore, we also found a phylogenetic link between these unclassified species and a CRESS virus DNA sequence previously identified in environmental wastewater in the United States in 2015. The findings, therefore, must be designated as exploratory to reveal potential novel species of CRESS-DNA viruses present in the vaginal tract of adult women. However, the potential for a pathogenic or commensal role of these novel CRESS-DNA viruses in the vagina, in association with either human cells or those of eukaryotic microbes, is sufficient to encourage further investigation. In addition, it is also important to acknowledge that further data are required to comprehensively grasp the taxonomy of CRESS-DNA viruses, given their significant diversity.
AUTHOR CONTRIBUTIONS
Conceptualization: Antonio Charlys da Costa, Iara M. Linhares, Steven S. Witkin and Steven D. Spandorfer. Methodology: Endrya do Socorro Foro Ramos, Tania Regina Tozetto-Mendoza, Pietro Bortoletto, Layla Honorato, Noely Evangelista Ferreira, Erick Matheus Garcia Barbosa, Adriana Luchs, Elcio Leal and Antonio Charlys da Costa. Formal Analysis: Endrya do Socorro Foro Ramos, Tania Regina Tozetto-Mendoza, Pietro Bortoletto, Layla Honorato, Noely Evangelista Ferreira, Erick Matheus Garcia Barbosa, Adriana Luchs, Elcio Leal and Antonio Charlys da Costa. Investigation: Endrya do Socorro Foro Ramos, Tania Regina Tozetto-Mendoza, Pietro Bortoletto, Layla Honorato, Noely Evangelista Ferreira, Erick Matheus Garcia Barbosa, Adriana Luchs, Iara M. Linhares, Steven D. Spandorfer, Elcio Leal, Antonio Charlys da Costa, Steven S. Witkin and Maria Cassia Mendes-Correa. Data curation: Tania Regina Tozetto-Mendoza, Elcio Leal and Antonio Charlys da Costa. Writing-original draft preparation: Endrya do Socorro Foro Ramos, Tania Regina Tozetto-Mendoza and Pietro Bortoletto. Writing-review and editing: Steven S. Witkin, Iara M. Linhares, Endrya do Socorro Foro Ramos, Tania Regina Tozetto-Mendoza and Pietro Bortoletto. Funding acquisition: Maria Cassia Mendes-Correa.
ACKNOWLEDGMENTS
ACdC is supported by a scholarship from HCFMUSP with funds donated under the #HCCOMVIDA scheme and LIM-52. This study was supported by Virology laboratory (LIM-52) with funds donated from Hospital das Clinicas of Faculdade de Medicina da Universidade de São Paulo (HC-FMUSP) under the #HCCOMVIDA scheme.
CONFLICT OF INTEREST STATEMENT
The authors declare conflicts of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




