The downregulation of Tapasin in dendritic cell regulates CD8+ T cell autophagy to hamper hepatitis B viral clearance in the induced pluripotent stem cell-derived hepatocyte organoid
Jinmei Chen and Leer Shen contributed equally to this study.
Abstract
Tapasin, a crucial molecular chaperone involved viral antigen processing and presentation, plays an important role in antivirus immunity. However, its impact on T cell differentiation in the context of virus clearance remains unclear. In this study, we employed induced pluripotent stem cells to differentiate into hepatocyte-like cell, which were subsequently inserted to the inverted colloidal crystal scaffolds, thus establishing a hepatocyte organoid (HO). By inoculating hepatitis B virus (HBV) particles in the system, we successfully engineered a robust in vitro HBV infection model for at least 3 weeks. Furthermore, we aimed to explore the effects of lentivirus-mediated short hairpin RNA (shRNA) targeting human Tapasin on the differentiation and antiviral function of CD8+ T cells. Specifically, we transfected dendritic cells (DCs) with Tapasin-shRNA and cocultured with T cells. The results demonstrated that Tapasin-shRNA transfected DCs effectively suppressed T cell proliferation and impeded HBV-specific cytotoxic T lymphocyte responses. Our investigation also revealed the role of mTOR pathway activation in reducing autophagy activity within CD8+ T cells. Expressions of autophagy-related proteins, beclin-1, LC3II/LC3I were decreased and PI3K/AKT/mTOR activity was increased in Tapasin-shRNA group. Collectively, our findings elucidate that shRNA targeting the Tapasin gene within DCs inhibits T cell differentiation by reducing autophagy activity to hamper viral clearance in the HBV-infected HO.
1 INTRODUCTION
Hepatitis B virus (HBV) infection is a leading cause of hepatocellular carcinoma and cirrhosis.1, 2 Current antiviral therapy only led to the elimination of HBV in a very small number of chronic hepatitis B (CHB) patients. HBV persists in part due to the steady loss of HBV-specific CD8+ T cells in CHB. HBV-specific CD8+ T cells are pivotal for the complete eradication of HBV infection. In healthy individuals, CD8+ T cells expand quickly and become effector cells with a focus on exerting an immediate effector role after activation, but this falls short in chronic HBV carriers.3 The restoration of HBV immunity, particularly HBV specific CD8+ T cells, is the goal of the development of new treatment approaches for CHB.
Tapasin plays a vital role in CD8+ T cell-mediated cellular immune responses. The viral antigen binded to the major histocompatibility complex-I (MHC-I) molecule is subsequently given to T cells as the resulting peptide loading complex. In this process, Tapasin completes a committed step by delivering antigenic peptides to MHC-I molecules in the endoplasmic reticulum and presenting them on the surface of antigen presenting cells for recognition by CD8+ T cell, thus catalyzing peptide loading of MHC-I.4 A previous study found that Tapasin-knockout mice showed decreased amounts of MHC-I, which had an impact on antigen presentation, CD8+ T cell maturation, and antiviral immunity.5 Our earlier studies confirmed that Tapasin modification can induce dendritic cell (DC) maturation and promoting the differentiation of HBV-specific CD8+ T cells.6
General experiment is designed to mimic internal environment, increasing the complexity of the in vitro hepatocyte culture system. It has tremendous potential applications in disease modeling and drug development. Most experimental animals do not express human sodium taurocholate cotransporting polypeptide (NTCP), making it impossible to accurately mimic the HBV infection process. NTCP-express cell lines can be vulnerable to HBV infection, but the whole course of HBV infection cannot be achieved.7 Induced pluripotent stem cell (iPSC) can differentiate into various cells, providing an alternative for limited organ supply.8 iPSC-hepatocyte-like cells (HLCs) can express mature hepatic cell markers, including NTCP, which is necessary to support HBV infection. Since it depicts the pertinent organ characteristics and the three-dimensional structure of human tissues, hepatocyte organoids (HOs) help us understand the biological system. Mechanistically, iPSC-derived hepatic cells are vulnerable to HBV and 3D cultured HOs have been proven to be more vulnerable to HBV infection than 2D normal liver cell culture.9
In our study, we established an HBV-infected HO successfully. Knockdown of Tapasin could cause inefficient antigen presentation function, thereby affecting T cell immune function. Then the antiviral efficacy of T cell was decreased in HBV-infected HO. The potential mechanisms might be associated with lower autophagic activity in CD8+ T cells, suggesting that chaperones-mediated MHC-I/peptide complex assembly was crucial for HBV immune clearance.
2 RESULTS
2.1 Establishment of HBV-infected HO
To establishing the HO for HBV disease modeling, iPSCs were differentiated into HLCs and then seeded into the inverted colloidal crystal (ICC) scaffold for further maturation (Figure 1A). HLCs were generated over a 21-day differentiation through the orchestrated application of growth factors and chemicals cocktails, stimulating the liver development.10 The differentiated HLCs were verified by the upregulation of hepatic cell markers and reduction in stem cell markers (Figure 1B–D). The transition from pluripotent states to hepatic phenotype was characterized by the downregulation of stem cell markers NANOG, OCT4, and SOX2, contrasting the upregulation of hepatic cell markers HNF4α, CYP3A4, ALB, and NTCP (Figure 1D–F).
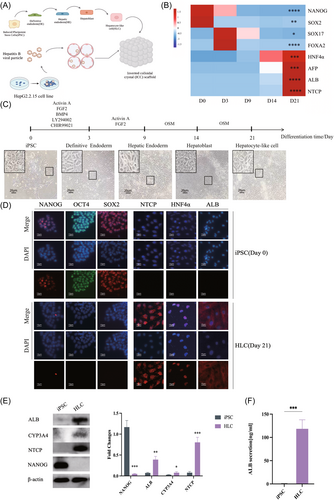
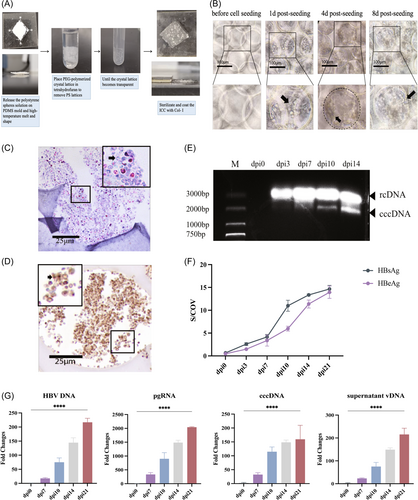
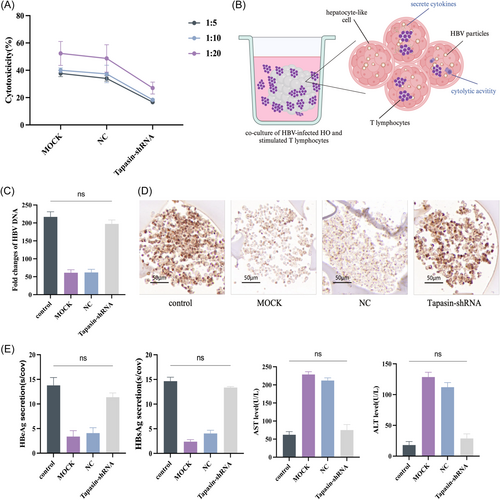
Upon achieving the HLC state, ICC scaffolds were employed to construct HO. The initial cell seeding resulted in the cell–extracellular matrix (ECM) surface interaction that progressively expanded toward the core, culminating to the formation of mature HO (Figure 2A–C). Subsequently, HBV particles were introduced to the HO at a concentration of multiple of infection (MOI) 300. On the 14th day postinfection, immunohistochemical analysis showed that HBsAg were expressed in the cytoplasm of ICC cells (Figure 2D). Notably, the existence of covalently closed circularDNA (cccDNA) was detected through DNA agar gel electrophoresis (Figure 2E). In addition, HBV had successfully infected the HO as evidenced by the expression of HBV DNA, pgRNA, cccDNA, supernatant vDNA, HBcAg, and HBeAg on the 7, 10, 14, and 21 days postinfection compared to the preinfection levels (Figure 2F,G).
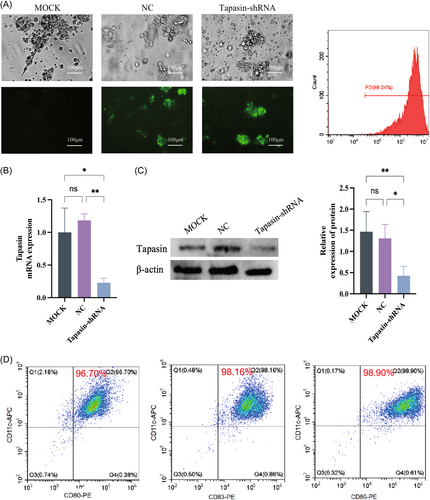
2.2 Tapasin-short hairpin RNA (shRNA) reduces the level of Tapasin in DCs and inhibits the secretion of cytokines in T cells
To reduce the expression of Tapasin in DCs, we developed the Tapasin-shRNA. Efficient transfection of the recombinant lentivirus plasmid into DCs was observed by green fluorescence detection using flow cytometry and fluorescent inverted microscopy (Figure 3A). Tapasin mRNA and protein levels exhibited markedly decrease after 72 h posttransfection (Figure 3B,C). Subsequently, the above DCs were induced maturation by HBcAg18–27 antigen, and assessed by the expression of cell surface molecules CD80, CD83, and CD86 as measured by flow cytometry (Figure 3D).
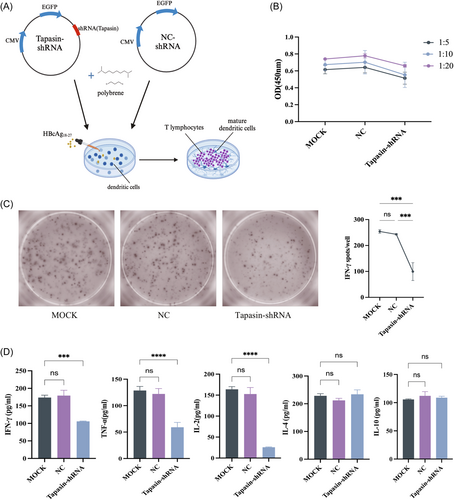
Upon coculturing DCs with T cells (Figure 4A), T cell proliferation was significantly lower upon stimulation by Tapasin-shRNA transfected DCs compared to the MOCK and NC groups across various cell ratios (1:5, 1:10, 1:20), indicating the influence of Tapasin downregulation on T cell proliferation (Figure 4B). The levels of IFN-γ, IL-4, TNF-α, IL-2, and IL-10 in the cocultured supernatant were also determined using ELISA and ELISPOT. IFN-γ, TNF-α, and IL-2 secretion levels significantly decreased, while IL-4 and IL-10 remained unchanged upon coculturing (Figure 4C,D). These outcomes suggested that Tapasin-shRNA intervention inhibited the secretion of cytotoxic T lymphocyte (CTL) cytokines IFN-γ and TNF-α, consequently affected their noncellular killing function.
2.3 Diminished Tapasin expression impairs the antiviral efficacy of T lymphocytes in the HO
The antiviral efficacy of T cells was assessed by Nonradioactive lactate dehydrogenase (LDH) release assays. The results revealed that HBV-specific CTL response was weakened upon the downregulation of Tapasin expression (Figure 5A). In contrast, there was no discernible difference in HBsAg, HBeAg, or HBV DNA in the Tapasin-shRNA group after coculturing the treated T cells with HBV-infected HO (Figure 5B), while the untreated group displayed gradual decline in HBsAg, HBeAg, and HBV DNA levels (Figure 5C–E). This observation suggests the impact of Tapasin downregulation on specific CTL response. Coincidentally, the destruction of cells led to increased release of AST and ALT (Figure 5E). Furthermore, immunohistochemical analysis of HBsAg in HO indicated comparable HBsAg levels between the Tapasin-shRNA group and the blank group, suggesting that T cells were unsuccessful in effectively eliminating HBV from the HO (Figure 5D).
2.4 Diminished Tapasin expression suppresses autophagy via PI3K/AKT/mTOR pathway in CD8+ T cells
To investigate the mechanism of Tapasin-shRNA-induced inhibiton of specific CTL response, we detected the levels of PI3K/AKT/mTOR pathway and their associated phosphorylated proteins in CD8+ T cells. Our findings showed that the PI3K/AKT phosphorylation level was reduced, while the mTOR phosphorylation level was increased (Figure 6A). It indicated the inhibition of specific CTL responses by Tapasin-shRNA may be related to the activation of the PI3K/AKT/mTOR pathway.
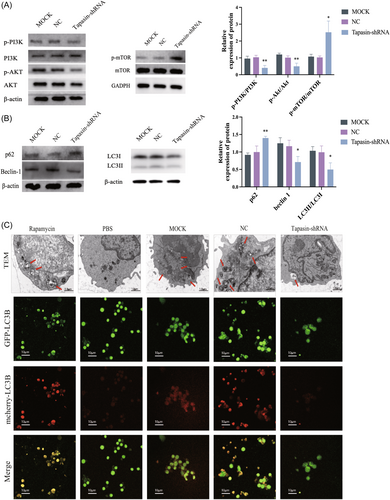
To further examine the precise effect on T cell differentiation, we conducted Western blot analysis to assess the expression of autophagy pathway related proteins (LC3, Beclin-1, and p62). Relative to the MOCK and NC groups, the Tapasin-shRNA group displayed reduced levels of both LC3II and LC3I, a decreased LC3II/LC3I ratio, and elevated p62 levels, and decreased Beclin-1 levels (Figure 6B). Moreover, transmission electron microscopy analysis unveiled a notably lower quantity of autophagosomes in the Tapasin-shRNA group compared to the MOCK and NC groups. In addition, T cells transfected with Ad-mCherry-GFP-LC3B were cocultured with DCs. The laser confocal microscopy analysis indicated more cells in the MOCK and NC groups expressed red and yellow fluorescence (activated autophagy), however, Tapasin-shRNA group where such fluorescence was subdued (Figure 6C). Therefore, Tapasin-shRNA transfection into DCs could effectively hinder T lymphocyte autophagy.
2.5 mTOR inhibition activates CD8+ T lymphocyte autophagy to suppress HBV replication
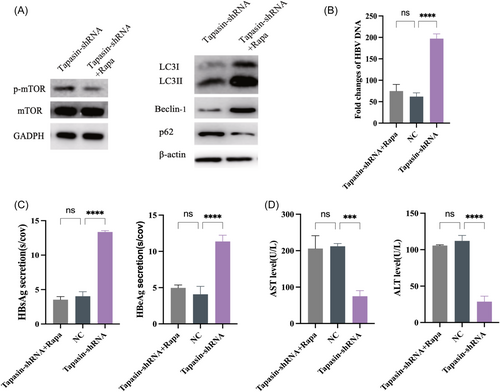
To validate the association of Tapasin with the mTOR pathway and autophagy in CD8+ T cells, we introduced the mTOR inhibitor rapamycin into Tapasin-shRNA group. Compared with Tapasin-shRNA group and NC group, the lower mTOR levels and higher LC3II/LC3I ratios were detected in Tapasin-shRNA + Rapa group. Simultaneously, the p62 level was downregulated while the Beclin-1 level was elevated. The results were indicated that the addition of autophagy inducers could activate CD8+ T cells autophagy in Tapasin-shRNA + Rapa group (Figure 7A). Consequently, the levels of ALT and AST were increased and the levels of HBV DNA, HBsAg, and HBeAg were decreased in Tapasin-shRNA + Rapa group (Figure 7B–D).
3 DISCUSSION
The scarcity of primary human hepatic cells, coupled with their challenges in expansion and phenotype preservation during in vitro culture, has prompted the exploration of alternative approaches. iPSCs have emerged as a promising solution, offering the potential for an inexhaustible supply of HLCs.11, 12 This study embraces the concept of mimicking human liver development by orchestrating the hepatic differentiation using specific culture conditions, guided by growth factors and chemical cues.10 The generation of HLCs was characterized by the expression of key hepatic genes such as ALB, CYP3A4, HNF4α, and NTCP.13
To further mature the HLC for HBV disease model development, we leverage the innovative ICC scaffolds constructed with polyethylene glycol (PEG). The unique design features of the ICC scaffold enable us to modulate the cell–cell, cell–ECM interactions and tissue connectivity, hence offer a novel platform for investigating host–pathogen interactions. Similar to our previous HCV liver infection disease modeling using ICC scaffold,10, 14 we successfully established an HBV-infected HO derived from iPSCs and HBV DNA, pgRNA, HBeAg, and HBsAg expression could be detected. However, the current system was only maintained a limited duration of HBV infection (just 21 days), which may be due to the gradual loss of the proliferative ability and hepatocyte-specific function of HLC in vitro over time. The urgent need is required to improve the differentiation method, extend infection duration and optimize effectiveness.
CD8+ T cell is known to play pivotal role in immune surveillance, employing both IFN-γ production and cytolytic activity to eliminate infected cells and tumors.15 Immunomodulation, a critical step in CHB management, is heavily influenced by DCs. Tapasin, acting as a molecular chaperone, contributes to antigen presentation in DCs. It aids in the peptide loading process, peptide-MHC I complex assembly, and endogenous antigenic peptide processing, thereby influencing CTL recognition of target cells.16 Enhanced antigen presentation and promotion of HBV-specific CD8+ T cells have been observed through Tapasin antigen modification in animal studies.17 Our study attempted to explore the mechanism of Tapasin for inducing T cell differentiation within the HBV-infected HO.
Studies have demonstrated that inflammatory cytokines (IFN-γ and TNF-α) released by CD8+ T cells exert control over HBV by inhibiting HBV replication, eliminating capsid, and disrupting the stability of cccDNA.18 Additionally, balanced cytokine levels, such as IFN-γ, IL-17A, and IL-21, contribute to effective T cell responses for infection clearance, while heightened IL-10 dampens these responses, leading to HBV chronicity.19 Our findings indicated that Tapasin-shRNA-transfected DCs curbed T cell proliferation and diminished Th1 cytokine secretion, particularly IFN-γ. Our previous group found that Tapasin overexpression increased the production of IFN-γ and IL-2.20, 21 This depletion in antigen presentation by DCs hindered efficient viral antigen delivering to CD8+ T cells and impeded HBV elimination in the HO (Figure 8).
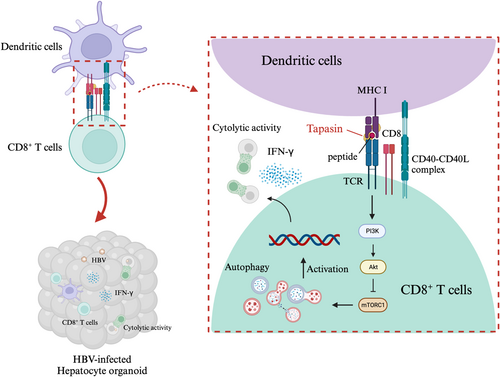
The mTOR pathway, closely associated with autophagy, plays a vital role in T cell metabolic reprogramming for survival. Our findings suggested that Tapasin-shRNA transfection into DCs might activate mTOR and inhibit autophagy in CD8+ T cells to consequently weaken specific HBV immune responses. It is notable that IFN-α2a treatment's downregulation of AKT/mTOR signaling augments autophagosome production by restraining autophagic degradation.22
Generally, after being stimulated by antigens, T cells must rapidly grow, mature, and carry out effector functions. Increased autophagy is one of the primary strategies to address the enormous need for bioenergy. Researchers have found that animals without autophagy have much fewer peripheral T cells, are more susceptible to apoptosis, and have damaged mitochondria that replicate at the same time, boosting ROS production and cell death.23 Besides that, autophagy plays a crucial role in the development and function of lymphocyte.24 In adaptive immunity, autophagy regulates how antigens are prepared and presented as well as how lymphocytes mature. When activated, their cell metabolism will dynamically transition from aerobic glycolysis to oxidative respiration.25 The maintenance of cytotoxic T cell clonal development requires this change, which makes CTL highly proliferative and produces inflammatory cytokines and cytolytic particles.26, 27 Autophagy is a constant intracellular volume degradation system that eliminates damaged cell components. It is essential for keeping lymphocytes' metabolic balance and survival in check.28
In summary, we successfully constructed an HBV-infected HO and indicated that the lack of Tapasin could reduce the immune function of HBV antigen-specific CD8+ T cells, which might cause by the modifications of the autophagy activity in CD8+ T cells. Further work needs to be done to evaluate precisely how Tapasin influences T cell activation and subsequent effector responses in treatment of CHB.
4 MATERIALS AND METHODS
4.1 iPSC and differentiation
iPSC (ZB11AN-B) was purchased from Shanghai East Hospital in China. Human umbilical cord blood monocytes were induced into iPSC by exogenous transcription factors mediated by Sendai virus. Cells were inoculated with the Essential 8 medium (Thermo Fisher Scientific) in a Petri dish coated with Laminin 521 (Bio-Lamina) until they reached 80% confluence. iPSC was differentiated using an established protocol.10 Briefly, endoderm formation was induced by Essential 6 medium supplemented with 3 μmol/L CHIR99021, 10 μmol/L LY294002, 10 μg/L BMP4, 80 μg/L FGF2, and 100 μg/L Activin A for 48 h. Subsequently, cells were cultured in RPMI-1640/1x B27 supplemented with 50 μg/L Activin A for 6 days to achieve hepatoblast formation. For hepatic maturation, cells were cultured in William's E medium (Gibco) in the presence of 20 μg/L OSM until 3 weeks (Supporting Information: Table 1). The chemicals and regents were purchased from PeproTech and Thermo Fisher Scientific.
4.2 HBV-infected HO
The establishment of the ICC scaffolds is based on the previously reported methods.10 Briefly, polystyrene microspheres were poured into the mold, melted and shaped at a high temperature. Then PEG diacrylate and photosensitive initiator were used to fill in the gaps and establish the scaffold's shape under UV radiation.
Tetrahydrofuran was used to dissolve the polystyrene microspheres and collagen I coated the scaffold. iPSC differentiated for 21 days and inoculate them into ICC scaffold to construct a HO. Meanwhile, the supernatant of HepG2.2.15 cells were collected and separate HBV particles with PEG8000 by centrifugal force. The virus titer of HBV DNA was detected by fluorescence quantitative polymerase chain reaction (PCR) diagnostic kit (Kanglang). The above virus particles were inoculated into the HO at the concentration of infection (MOI) 300. The William's E medium (Gibco) containing 4% PEG8000 and 2% dimethyl sulfoxide (DMSO) was used to incubate for 24 h. Phosphate buffered saline (PBS) was washed three times. The medium without PEG8000 was replaced for further culture for 3 weeks. On the 0, 7, 10, 14, and 21 days postinfection, cells and supernatant were collected for PCR and ELISA, respectively. Immunohistochemical staining of HBsAg was performed on the 14 days postinfection, and cccDNA electrophoresis was performed on the 3, 7, 10, and 14 days postinfection.
4.3 Plasmid construction, identification and transfection
In accordance with a published sequence of human Tapasin (NM 003190.5), certain shRNAs specifically targeting human Tapasin were designed to knockdown its expression. The shRNA sequences are listed in Supporting Information: Table 2. They were annealed for 5 min in a buffer that was heated to 95°Cand then cooled down to room temperature. At the EcoRI and AgeI sites, the generated double-stranded oligo DNA was cloned into plasmid. In addition, enhanced green fluorescent protein was expressed by the pSLenti-U6-shRNA-CMV-EGFP-F2A-Puro-WPRE.The above plasmids, Tapasin-shRNA and NC-shRNA, were transformed into competent cells (DH5α; Biowit Technologies), and extracted with a plasmid purification kit. Successful ligation was determined by PCR and sequencing analyses. The recombinant expressing Tapasin or NC shRNA vectors were next transfected into 293 T cells and measured. DCs were seeded on a 12-well plate at a density of 3 × 105 cells/well. Lentiviral stocks were diluted with serum-free RPMI-1640 medium to MOI = 100, then added to the DCs culture with 5 μg/mL polybrenes to incubate for 24 h at 37°C. The culture medium was then replaced with fresh RPMI-1640 complete medium for another 48 h. The efficiency of knocking down Tapasin was subsequently evaluated by Real time PCR and Western Blot.
4.4 Culture of DCs and T cells
PBMC was purchased from Shanghai Jiayuan Biological Co., Ltd. T cells were isolated using the nylon filtration method, and cultured (2 × 106 cells/mL) in complete RPMI-1640 in the presence of 2 μg/mL CD28 antibody and 50 ng/mL IL-2. DC was isolated using the adhesion method and cultured (106 cells/mL) in complete RPMI-1640 in the presence of 100 ng/mL IL-4 and 100 ng/mL GM-CSF at 37°C. On the 5th day, Tapasin-shRNA was transfected into DCs. Then cells were exposed to 10 μg/mL HBcAg18-27 (GL Biochem) and 75 ng/mL TNF-α on the eighth and ninth days, respectively. On the 10th day, DC and T cells were cocultured in a 1:10 ratio for 72 h. After DC stimulation, T cells (5 × 106 cells/HO) were co cultured with HBV infected HO 14 days postinfection for 72 h. HO cells and supernatant were collected for HBV DNA and HBsAg, HBeAg, ALT, AST detection, respectively. Immunohistochemical staining of HBsAg was performed. All cytokines were purchased from PeproTech.
4.5 Flow cytometry and FACS
Cells were resuspended in PBS and cell suspensions were stained with PE-conjugated human antibody (described in Supporting Information: Table 3) for 30 min on ice. Analysis was performed by flow cytometry (Thermo Fisher Scientific).
T cells were resuspended in PBS and stained with APC-conjugated human antibody (described in Supporting Information: Table 3) for 30 min. CD8+ T cells were separated by Fluorescence Activated Cell Sorter (Thermo Fisher Scientific).
4.6 Cell counting kit-8 (CCK-8) assay
On the respective conditions of the DC: T cells ratio as 1:5, 1:10, and 1:20, cells were cocultured in 96-well plates for 24 h at 37°C with 5% CO2. CCK-8 solution (Dojindo) was added and incubated for 2 h. The value of OD was measured at 450 nm in a microplate reader (Thermo Fisher Scientific).
4.7 ELISA and ELISPOT
The levels of HBsAg and HBeAg in the coculture supernatants were measured by commercially available ELISA assays (Kehua Biotech Co., Ltd.). The levels of IFN-γ, TNF-α, IL-10, IL-4, and IL-2 in the coculture supernatants of effector and target cells were assessed by commercially available ELISA assays (Dakewe Biotech Co., Ltd.). The levels of IFN-γ in the coculture supernatants were assessed by commercially available IFN-γ ELISPOT assays (Dakewe Biotech Co., Ltd.). AST/ALT levels were detected by commercially available AST/ALT Assay Kit (Nanjing Jiancheng Bioengineering Institute).
4.8 LDH release assay
CD8+ T cells as the effector cells and HepG2.2.15 cells as target cells were cocultured in 96-well plates at different effector-to-target (E/T) ratios (5:1, 10:1, and 20:1) at 37°C with 5% CO2 for 4 h. The killing efficiency of CIK was detected by LDH releasing assay (CytoTox 96 Non-Radioactive Cytotoxicity kit; Promega). The absorbance values of the supernatants were recorded at optical density 490 nm. Cytotoxicity (%) was calculated as follows: [(experimental release − effector spontaneous release − target spontaneous release)/(target maximum release − target spontaneous release)] × 100%.
4.9 Transmission electron microscopy
The purified CD8+ T cells in groups were washed with PBS for three times, and treated with triptolide for 24 h. Then, the cells were fixed with 4% paraformaldehyde at 4°C for 1.5 h, washed at room temperature with 1% phosphate-buffered osmium tetroxide and dehydrated gradually with ethanol. The dehydrated pellets were washed with propylene oxide at room temperature for 30 min. After embedding and double staining with uranyl acetate/lead citrate, the sections were determined by transmission electron microscopy (Hitachi).
4.10 Immunofluorescence
Cells were adhered to poly-L-lysine-coated cover slips, fixed with 2% paraformaldehyde, permeabilized with 0.1% Triton X-100 and stained with corresponding primary and secondary antibodies (described in Supporting Information: Table 4). At last, cells were counterstained with Hoechst 33342 nuclear dye.
4.11 Immunohistochemistry
The HO was fixed with formaldehyde and embedded in paraffin. For immunohistochemistry, section 3–5 mm thick were treated with 0.3% H2O2 for 10 min to inactivate endogenous peroxidase, and 2% normal goat serum was blocked for 30 min at room temperature. Then, goat anti-HBsAg were added overnight. After washing three times with PBS, the sections were stained with biotinylated secondary antibody (Chinese Post) at 37°C for 30 min and incubated with streptavidin biotin peroxidase complex for 30 min. The visualization was carried out with diaminobenzidine (booster) and haematoxylin.
4.12 Real time PCR
According to the manufacturer's protocol, total RNA was extracted using TRIZOL reagent and quantified with NanoDrop. cDNA was synthesized using 500 ng of total RNA with a Superscript VILO cDNA synthesis kit (Thermo Fisher Scientific). Real-time PCR was performed with SYBR Green PCR Master Mix (Thermo Fisher Scientific) using a StepOnePlus real-time PCR system (Thermo Fisher Scientific). The relative quantitation of target mRNA levels was performed by using the 2−ΔΔCT method. The values were normalized by those of the housekeeping gene, glyceraldehyde 3-phosphate dehydrogenase. Primer sequences are listed in Supporting Information: Table 2.
4.13 Western blot
Cells were harvest to isolate the total protein with RIPA lysis buffer (Beyotime Institute of Biotechnology). Equal amounts of proteins from each sample were loaded onto polyacrylamide mini gels (Invitrogen) for electrophoresis. Proteins from the gels were transferred to Immobilon-PVDF membranes (Millipore) using a semidry apparatus (BioRad). After incubation with corresponding antibodies (described in Supporting Information: Table 5), the signals were detected with an ECL assay kit (Amersham).
4.14 Southern blot
For the detection of cccDNA, a modified Hirt method was used to extract protein-free vial DNAs as described. Briefly, protein-free vial DNAs were isolated from full six-well plate HBV infected HLCs. The extracted DNA was treated with EcoR I overnight. The viral DNAs were separated on agarose gel, transferred to an Amersham Hybound-N+ membrane (GE Healthcare). HBV-specific digoxigenin-labeled prime was generated and detected with DIG DNA labeling and detection Kit (Roche).
4.15 Statistical analysis
The analysis was pooled from n = 3 independent experiments. Mean and standard deviation (SD) from independently repeated experiments were plotted and subjected to unpaired two-tailed t-tests (Student's t-test) or one-way ANOVA. p Value < 0.05 was considered statistically significant. GraphPad 9 was used for plotting and statistical testing.
AUTHOR CONTRIBUTIONS
Jinmei Chen and Leer Shen contributed equally to this study, including the experimental implementation and data collation, as well as the writing and submission of the article. Qingxin Guo and Siyuan Ma participated in experimental data collation and article writing. Yi Zhang and Jie Chen participated in the experiment implementation and data collation. Lihong Qu, Soon Seng Ng, and Xiaohua Chen participated in experimental design, data collation, article writing, revision, and submission.
ACKNOWLEDGMENTS
The financial supports were from the National Natural Science Foundation of China (82070615) and Shanghai Science and Technology Commssion (21140901100).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




