SARS-CoV-2 infection triggers more potent antibody-dependent cellular cytotoxicity (ADCC) responses than mRNA-, vector-, and inactivated virus-based COVID-19 vaccines
Abstract
Neutralizing antibodies (NAbs) are elicited after infection and vaccination and have been well studied. However, their antibody-dependent cellular cytotoxicity (ADCC) functionality is still poorly characterized. Here, we investigated ADCC activity in convalescent sera from infected patients with wild-type (WT) severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) or omicron variant compared with three coronavirus disease 2019 (COVID-19) vaccine platforms and postvaccination breakthrough infection (BTI). We analyzed ADCC activity targeting SARS-CoV-2 spike (S) and nucleocapsid (N) proteins in convalescent sera following WT SARS-CoV-2-infection (n = 91), including symptomatic and asymptomatic infections, omicron-infection (n = 8), COVID-19 vaccination with messenger RNA- (mRNA)- (BNT162b2 or mRNA-1273, n = 77), adenovirus vector- (n = 41), and inactivated virus- (n = 46) based vaccines, as well as post-mRNA vaccination BTI caused by omicron (n = 28). Correlations between ADCC, binding, and NAb titers were reported. ADCC was elicited within the first month postinfection and -vaccination and remained detectable for ≥3 months. WT-infected symptomatic patients had higher S-specific ADCC levels than asymptomatic and vaccinated individuals. Also, no difference in N-specific ADCC activity was seen between symptomatic and asymptomatic patients, but the levels were higher than the inactivated vaccine. Notably, omicron infection showed reduced overall ADCC activity compared to WT SARS-CoV-2 infection. Although post-mRNA vaccination BTI elicited high levels of binding and NAbs, ADCC activity was significantly reduced. Also, there was no difference in ADCC levels across the four vaccines, although NAbs and binding antibody titers were significantly higher in mRNA-vaccinated individuals. All evaluated vaccine platforms are inferior in inducing ADCC compared to natural infection with WT SARS-CoV-2. The inactivated virus-based vaccine can induce N-specific ADCC activity, but its relevance to clinical outcomes requires further investigation. Our data suggest that ADCC could be used to estimate the extra-neutralization level against COVID-19 and provides evidence that vaccination should focus on other Fc-effector functions besides NAbs. Also, the decreased susceptibility of the omicron variant to ADCC offers valuable guidance for forthcoming efforts to identify the specific targets of antibodies facilitating ADCC.
1 INTRODUCTION
The coronavirus disease 2019 (COVID-19) pandemic, caused by the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), continues to spread, resulting in more than 651 million cases and 6.9 million deaths as of April 30, 2023.1 Since the declaration of COVID-19 as a pandemic by the World Health Organization, the world has entered a state of alert, and several effective vaccines have been rapidly developed and deployed globally.2 These vaccines and other antiviral interventions were designed against the original SARS-CoV-2 strain that emerged in 2019 in Wuhan, China.3 Since then, several variants emerged that are more transmissible and capable of escaping both vaccine- and infection-induced immunity. Although some studies have shown that vaccines can still induce an immune response against these variants, they are not as effective against emerging variants.4, 5 This has dampened the progress in controlling the pandemic, creating concerns and raising questions regarding the future evolutionary trajectories of SARS-CoV-2 variants. Hence, the challenge remains to identify new immune correlates of protection following SARS-CoV-2 infection or COVID-19 vaccination.
The current knowledge regarding the mechanisms by which antibodies can protect against COVID-19 infection and disease or, conversely, contribute to disease development has been limited to neutralizing activity and cellular cytotoxicity. In COVID-19 studies, interferon responses were the main subject of study during the innate phase of infection, whereas neutralizing activity and T-cell responses were investigated during the adaptive phase.6 Immune responses against SARS-CoV-2 spike (S) protein, particularly neutralizing antibodies (NAbs), essentially contribute to the protection following vaccination and natural infection.7 These NAbs are detectable within the second week following disease onset in 94% of recovered COVID-19 patients.8 However, the NAbs titers vary highly across studies and wane about 6 months after infection.
Besides neutralization, S-specific antibodies can eliminate viruses or virus-infected cells via other mechanisms, including antibody-dependent cellular cytotoxicity (ADCC), antibody-dependent cellular phagocytosis (ADCP), or complement-dependent cytotoxicity.9, 10 There has been considerable interest in the ADCC immune mechanism through which Fc receptor-bearing immune cells can recognize and kill antibody-coated target cells expressing tumor- or pathogen-derived antigens on their surface.11 Effector immune cells, most commonly natural killer (NK) cells, bind to the antibodies through their CD16 receptors (FcγRIII). CD16-mediated activation of NK cells results in degranulation by releasing cytotoxic molecules such as perforin and granzyme.12
Despite the importance of NK cells in providing immunity against viral infections and bridging the innate and adaptive immune responses, they have been investigated in a limited number of COVID-19 studies.13 In animal models, Fc-mediated effector functions were shown to correlate with protection.14 Also, antibodies mediating ex vivo ADCC activity against the S protein were shown in convalescent plasma from recovered COVID-19 patients.15 Here, we aimed to investigate ADCC responses induced by natural infection and identify the extent to which different COVID-19 vaccines can induce NK-cell–mediated ADCC and whether it differs from natural infection. This is the first study to evaluate ADCC effector function in fully vaccinated subjects with the adenovirus vector- and inactivated virus-based vaccines in comparison to messenger RNA (mRNA) vaccines and natural infection with wild-type (WT) SARS-CoV-2 and Omicron variant.
2 METHODS
2.1 Ethical approval and sample collection
A total of 291 samples were collected from SARS-CoV-2-infected patients during the period of SARS-CoV-2 WT predominance (April–August 2020, n = 91) and Omicron variant predominance (n = 36) and individuals fully vaccinated with COVID-19 mRNA vaccines (Pfizer, BNT162b2, n = 40; Moderna, mRNA-1273, n = 37), adenovirus vector-based vaccine (AstraZeneca, ChAdOx1-nCoV-19, n = 41), and inactivated virus vaccine (Sinopharm, BBIBP-CorV, n = 46). For SARS-CoV-2 infected patients, samples were collected within 2 weeks to 6 months postsymptoms onset for symptomatic patients or positive SARS-CoV-2 RT-PCR test for asymptomatic patients. Samples from COVID-19-vaccinated individuals were also collected within 2 weeks to 6 months postfull vaccination (second dose administration). For the control group, 20 prepandemic samples collected from healthy blood donors before 2019 were used to assess non-specific ADCC activity.16-18 Demographic and clinical data of the patients are shown in Table S1. Ethical approvals for sample collection were obtained from Qatar University (QU) institutional review board (QU-IRB # QU-IRB 804-E/17 and QU-IRB 1537-FBA/21), HMC (HMC-IRB# MRC-05-007), and the Primary Health Care Corporation's Independent Review Board (Ref. No. PHCCDCR202005047).
2.2 Serology testing for anti-SARS-CoV-2 antibodies
All sera samples were initially tested using CL-900i® automated analyzer (Mindray Bio-Medical Electronics),19 which detects antibodies targeting SARS-CoV-2 antigens including (i) anti-SARS-CoV-2 nucleocapsid (N) and spike (S) antigens immunoglobulin G (IgG), (ii) anti-S-RBD total antibodies (IgG, IgA, and IgM), and (iii) anti-S-RBD IgG. Anti-SARS-CoV-2 S1 IgA was measured using the Euroimmun anti-SARS-CoV-2 IgA enzyme-linked immunosorbent assay.20 All tests were carried out according to the manufacturer's instructions.
2.3 Neutralization assays
2.3.1 Pseudovirus production, titration, and neutralization test (pseudovirus neutralization test [pVNT])
A pVNT was used to measure NAbs against SARS-CoV-2 in vitro. The pseudovirus expresses the S glycoprotein, which mediates entry into the host cells by binding to the human receptor. Pseudoviruses expressing WT SARS-CoV-2 (Wuhan-Hu-1) and MERS-CoV S were prepared using human embryonic kidney (HEK293T) cells (American Type Culture Collection) infected with vesicular stomatitis virus ΔG-luc seed virus as previously described.21 For the pseudovirus titration assay, HEK293T cells were used, as described by Wang et al.22 Cells and plasmids were kindly provided by the Viral Pathogenesis Laboratory, Vaccine Research Center, National Institute of Health (NIH). Briefly, HEK293T cells were plated at 1 × 104 cells per well in a 96-well white/black isoplate (PerkinElmer) and cultured overnight. The following day, the culture medium was removed, and twofold serial dilutions of the pseudovirus were added to the cells and incubated for 2 h. Then, 100 μL of fresh medium was added, and after 72 h, cells were lysed using 1x lysis buffer, and 50 μL of luciferase substrate (Bio-Glo™ Luciferase Assay System, Promega) was added to each well. Luciferase activity was measured using a luminescence plate reader (Infinite pro200, Tecan). Following pseudovirus titration, the neutralization assay was performed as described elsewhere.22 Luciferase activity was measured using a luminescence plate reader (Tecan), and percent inhibition (%) was calculated for each sample.
2.3.2 Surrogate virus neutralization test (sVNT)
An automated competitive binding immuno-enzymatic assay (anti-SARS-CoV-2 NTAb assay) was used for quantitative detection of NAbs against SARS-CoV-2 RBD (Cat. No. SARS-CoV-2 Neutralizing Antibody 121, Mindray) for quantitative detection of nAbs against SARS-CoV-2 RBD. In this assay, anti-SARS-CoV-2 NAbs in the sample compete with ACE2-ALP conjugate for RBD-binding sites.23 The resulting chemiluminescent reaction is measured as relative light units (RLUs) by a photomultiplier built into the system, and the level of nAbs is determined via a calibration curve with a cutoff index of ≥10–1000 IU/mL.
2.4 ADCC assay
ADCC-mediating antibodies in plasma were measured using a commercial ADCC reporter bioassay kit (Promega).11 This assay utilizes ADCC effector cells (Jurkat-FcγRIIIa-NFAT-Luc, V158 high-affinity variant) to measure the ability of serum antibodies to activate the NFAT (nuclear factor of activated T cells) pathway through FcγRIII (the pathway that initiates ADCC in NK cells) in the presence of target antigens coated on a 96-well plate. ADCC was assessed against soluble prefusion stabilized His-tagged SARS-CoV-2 Spike S1 + S2 ECD (SinoBiological, Cat No. 40589-V08H4), SARS-CoV-2 B.1.1.529 (Omicron) Spike S1 + S2 protein (SinoBiological, Cat No. 50-222-1641) and, SARS-CoV-2 His-tagged full-length N protein (SinoBiological, Cat No. 40588-V08B). Briefly, 96-well plates were coated overnight at 4°C with 300 ng/well of S and 100 ng/well of N proteins diluted in 1x phosphate-buffered saline. Wells were then washed and blocked with 5% bovine serum albumin, and then serially diluted sera (in blocking buffer) heat-inactivated were added and incubated at 37°C for 2 h. ADCC effector cells were then added to each well (75 000 cells/well) and incubated overnight at 37°C. After incubation, a luciferase reagent (Bio-Glo) was used to measure luminescence activity using a luminescence plate reader (Tecan). ADCC activity was reported as fold change in the RLUs, which was calculated as follows: Fold of induction = RLU of induced/RLU of no serum control.
2.5 Statistical analysis
Statistical analysis was performed using GraphPad Prism 9.2.0. and Microsoft Excel 2013. Results were plotted as median values with the interquartile range. The difference between groups was evaluated using non-parametric one-way analysis of variance, and p ≤ 0.05 were considered statistically significant. Correlation and linear regression analysis between log-transformed assay readings were performed, and the Spearman correlation coefficient was calculated with a 95% confidence interval. Correlation interpretation was as follows: a coefficient of 0–0.39 indicates a weak correlation, 0.40–0.59 indicates a moderate correlation, 0.6–0.79 indicates a strong correlation, and 0.8–1 indicates a very strong correlation. In all graphs, significance was *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, or ****p ≤ 0.0001.
3 RESULTS
3.1 Anti-SARS-CoV-2 binding and neutralizing antibody responses
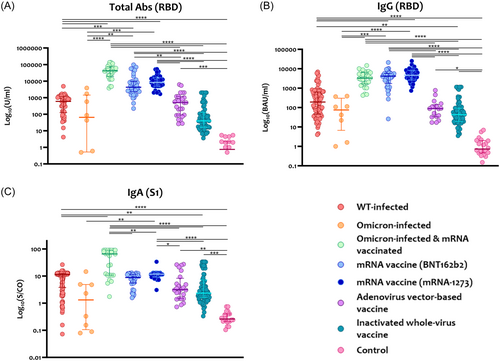
Antigen-specific serum anti-SARS-CoV-2 antibodies, including total immunoglobulins (Ig), IgG, and IgA, were measured using an automated immunoassay for all samples. As shown in Figure 1, binding antibodies, including total and IgG antibodies, were significantly higher in people vaccinated with COVID-19 mRNA vaccines (BNT162b2 and mRNA-1273) compared to those with naïve SARS-CoV-2 infection (WT or Omicron) or vaccinated with adenovirus vector and inactivated virus vaccines (Figure 1A,B). Further, post-mRNA vaccination breakthrough infection (BTI) showed significantly boosted levels of anti-RBD total Ig and IgG antibodies compared with naïve omicron infection (p < 0.001). The levels of anti-S1 IgA were variable between groups, but people infected with omicron and vaccinated with vaccines showed the highest levels compared to the other groups (Figure 1C).
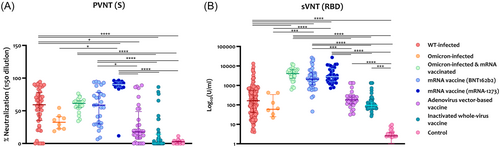
NAbs were measured using pVNT utilizing a pseudovirus expressing SARS-CoV-2 S protein and a sVNT measuring NAbs targeting the RBD. As shown in Figure 2A, the mRNA vaccine mRNA-1273 induced the highest NAbs against the S protein compared to natural infection and the other COVID-19 vaccines. Similarly, the BNT162b2 mRNA vaccine also induced significantly higher anti-S NAbs than the ones induced by the adenovirus- and inactivated virus-based vaccines. Adenovirus- and inactivated virus-based vaccines induced the lowest NAbs compared to natural SARS-CoV-2 infection and other COVID-19 vaccines. NAbs targeting the RBD also showed similar findings where mRNA vaccines induced the highest levels compared to natural infection (SARS-CoV-2 WT or Omicron variant) and adenovirus- and inactivated virus-based vaccines (Figure 2B). The levels of NAbs targeting the RBD were also higher in those with omicron BTI compared with naïve infection and adenovirus- and inactivated virus-based vaccines.
Neutralization of SARS-CoV-2 omicron was also assessed in samples collected from naïve omicron-infected people and those with omicron BTI (Figure S1A). Notably, in omicron naïve-infected patients, lower neutralization of omicron PV was observed compared to WT SARS-CoV-2, whereas BTI showed comparable neutralization levels of the WT and omicron.
3.2 SARS-CoV-2 spike (S) specific ADCC activity following infection and vaccination
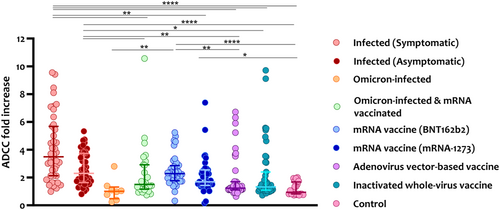
We evaluated ADCC activity induced by SARS-CoV-2 WT, omicron variant infections, and COVID-19 vaccination against SARS-CoV-2 whole S protein (WT). As shown in Figure 3, symptomatic patients induced the highest ADCC activity, reaching a 10-fold increase (average = 4.1, range = 1.0–9.6) in the assay signal. Asymptomatic patients (average = 2.6, range = 0.8–5.3) and BNT162b2-vaccinated individuals (average = 2.5, range = 0.3–5.2) also induced higher ADCC levels compared with omicron-infected patients and people vaccinated with the adenovirus- and inactivated virus-based vaccines. ADCC activity induced by naïve omicron infection (average = 1.1-folds), omicron BTI (average = 2.2-folds), mRNA-1273 vaccine (average = 2.1-folds), Adenovirus-based (average = 1.9-folds), and inactivated virus-based vaccines (average = 2.2-folds) showed comparable ADCC activity levels with no significant difference between them. Prepandemic controls did not induce any ADCC activity, showing less than a twofold increase (average = 1.6).
ADCC against SARS-CoV-2 omicron S protein was also assessed in samples from naïve omicron-infected people and those with omicron BTI. Interestingly, omicron-induced ADCC responses were reduced in the collected convalescent sera, ranging between 1.0 and 1.4-folds (average = 1.1) in naïve-infected people and between 1.0 and 2.6-folds (average = 1.3) in people with omicron BTI (Figure S1B).
We then assessed ADCC activity in high ADCC responders (ADCC fold increase ≥2) and obtained similar findings. Symptomatic patients showed the highest levels of anti-S ADCC activity and number of ADCC responders compared with mRNA-vaccinated groups (Figure S2A). Also, more symptomatic and asymptomatic patients were among the high ADCC responders compared with omicron-infected and COVID-19-vaccinated individuals.
3.3 SARS-CoV-2 nucleocapsid (N) specific ADCC activity following infection and vaccination
Since natural SARS-CoV-2 infection (WT or omicron variant) and the inactivated virus-based vaccine (Sinopharm) can induce immune responses to all SARS-CoV-2 antigens, we assessed whether they could induce ADCC activity against the N protein. As shown in Figure 4, natural WT SARS-CoV-2 infection induced significantly higher ADCC responses against the N protein ranging between 1.1 and 19.1-fold (average = 5.8) for symptomatic patients and 1.1–29.9-fold (average = 4.5) for asymptomatic patients compared with naïve omicron infection (average = 1.2), omicron BTI (average = 1.1), inactivated virus-based vaccine (average = 1.9) and prepandemic controls.
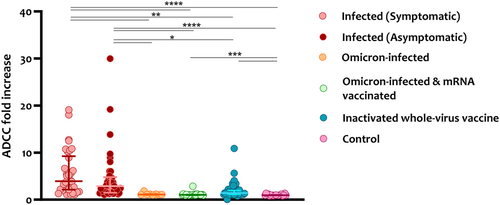
When assessing ADCC activity in high ADCC responders, no significant difference was observed across all groups except for the symptomatic patients who showed slightly higher levels than the inactivated virus-based vaccine (Figure S2B). Similarly, more symptomatic and asymptomatic patients were among the high ADCC responders.
3.4 ADCC activity over time
To assess the levels of ADCC activity over time, all sampling time points from each group were combined for analysis. ADCC activity was assessed following the onset of symptoms for infected patients or second dose administration for vaccinated individuals. Significant ADCC responses against the S protein were induced within the first month of symptom onset in all groups and remained detectable at comparable levels up to 6 months postsampling (Figure 5A–D).

We also assessed ADCC responses targeting the N protein over time in infected patients and people vaccinated with inactivated virus-based vaccines. We observed that ADCC activity was detectable within the first month of sampling and remained at comparable levels at all time points with no significant difference in both groups (Figure 5E,F).
Differences in ADCC activity in relation to age and gender were also evaluated. As shown in Figure S3, males showed higher ADCC responses against the S and N proteins in all assessed samples (Figure S3A,B; p = 0.0015 and p = 0.0172, respectively), but here was no significant difference when we separately assessed ADCC in the infected (Figure S3C,D) and vaccinated group (Figure S3E). Also, overall ADCC activity across the different age groups showed no significant difference (Figure S4A,B). However, ADCC levels against the S protein were higher in individuals aged 50–60 years than those between 19 and 40 years in the infected group (Figure S4C; p = 0.0015).
3.5 Correlation between ADCC and antigen-specific antibodies
We assessed the correlation between the levels of ADCC activity and antigen-specific antibodies, including binding and NAbs. Correlation and linear regression analysis were performed for infected SARS-CoV-2 patients and mRNA-vaccinated individuals since we have more samples from these groups than the other groups.
As shown in Figure 6A,B, the correlation between S-specific ADCC and IgG antibodies targeting both S and N proteins showed a moderate correlation in infected patients (r = 0.62, p < 0.0001) and a weak correlation in the mRNA-vaccinated group (r = 0.28, p = 0.027).
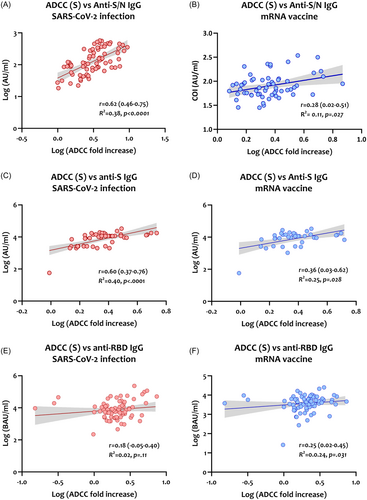
Similarly, S-specific ADCC activity showed a moderate correlation with anti-S IgG in infected patients (r = 0.60, p < 0.0001, Figure 6C) and a weak correlation in the mRNA-vaccinated group (r = 0.36, p = 0.028, Figure 6D). A poor correlation between ADCC activity and anti-RBD IgG in both infected patients (r = 0.18, p = 0.11, Figure 6E) and mRNA-vaccinated individuals (r = 0.25, p = 0.031, Figure 6F).
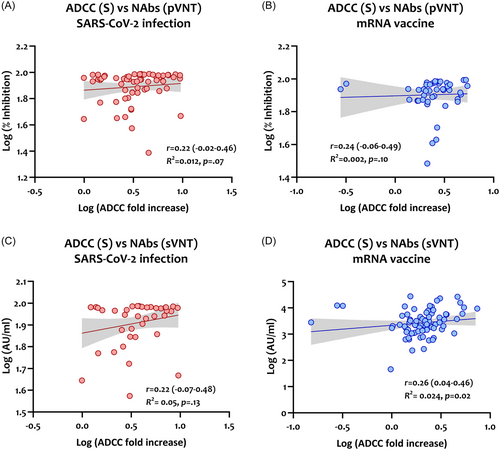
The levels of ADCC activity against the S protein were also correlated with NAbs targeting SARS-Cov-2 S protein and RBD, detected by pVNT and sVNT, respectively. As shown in Figure 7, S-specific ADCC activity correlated poorly with NAbs detected by the pVNT (r = 0.22 for infected patients and r = 0.24 for mRNA-vaccinated) and sVNT (r = 0.22 for infected patients and r = 0.26 for mRNA-vaccinated).
4 DISCUSSION
While NAbs are critical in impeding viral infections, other non-neutralizing immune Fc-mediated activities may hinder infection and limit disease severity.24 Fc-dependent ADCC effector functions were previously shown to play a role in the protection and pathogenesis of various infectious diseases.25 In terms of COVID-19, the exact role of ADCC is not yet fully understood, with only a few studies investigating Fc-mediated effector functions following infection and vaccination. In the present study, we analyzed binding, neutralization, and ADCC responses induced by natural infection with SARS-CoV-2 WT and omicron variant compared with four different COVID-19 vaccines and postvaccination BTIs caused by omicron infection. The assessed vaccine platforms are mRNA- (BNT162b1 and mRNA-1273), adenovirus vector-, and inactivated virus-based vaccines. ADCC activity was measured to evaluate SARS-CoV-2 antibodies with Fc effector function activity. This is the first study to assess ADCC effector function in sera samples collected from fully vaccinated subjects with adenovirus and inactivated virus vaccines compared to mRNA vaccines and natural SARS-CoV-2 infection.
Although natural SARS-CoV-2 infection elicited lower binding and NAbs than mRNA vaccines, it augmented higher NK-cell-mediated ADCC responses against SARS-CoV-2 S and N antigens. Further, symptomatic patients showed even stronger ADCC responses than asymptomatic and vaccinated individuals, particularly against the S protein. Previous reports indicated that during viral infection, elevated ADCC responses could induce inflammation,26 suggesting a potential role for ADCC in COVID-19 pathogenesis. Similar findings were reported in other studies which observed increased ADCC activity in severe disease.27 However, it was also shown that patients who survived severe disease induced higher ADCC levels than deceased patients, indicating that ADCC might have a protective role.28
On the other hand, elevated ADCC response in severe patients could result from prolonged SARS-CoV-2 infection with high viral load, leading to persistent antigen exposure in these patients.27 However, the timely induction of ADCC might play a role in determining the clinical outcome. Previous studies reported that the delayed production of NAbs correlated with severe COVID-19 and mortality.29 Hence, a similar mechanism might be involved in ADCC, in which the timing of antibody production plays a critical role in determining the clinical outcome. That being said, patients who develop delayed ADCC response might succumb to more severe disease. A possible explanation is that during the early stages of the disease, cells mediating ADCC might become functionally compromised and thus deprive the host of inducing ADCC responses. A study by Krämer et al.30 showed that severe COVID-19 was associated with impaired NK-cell activity and function. However, the impact of this impairment on ADCC responses in these patients is still unclear and needs further investigation.
In addition, we found that peak levels of S-specific ADCC responses were reached around 10–20 days, which was earlier than the reported peak time of NAbs, about 3 weeks after disease development.27, 29 Further, ADCC activity remained detectable for 6 months after disease onset and vaccination. A recent study assessing S-specific ADCC-mediating antibodies also showed early detection of ADCC responses within 10 days postinfection, which remained at relatively stable levels for up to 1 year.27 This extends the window beyond that inferred from the neutralizing activity and supports the possible application of ADCC as a prominent marker for disease prognosis. Still, this needs to be confirmed by testing more samples collected over extended time points while considering various vaccination and infection status combinations.
In line with other studies, our findings showed that vaccination induces lower ADCC responses than infection with WT SARS-CoV-2.27, 31 Although mRNA vaccination induces robust NAb titers following the second vaccination,32 they were less effective in inducing ADCC compared with natural infection. Also, no difference in ADCC levels was observed across the four COVID-19 vaccine platforms. These observations could partially explain the less effective protection provided by COVID-19 vaccines against infection and symptomatic diseases compared to natural infection-induced immunity. Increasing evidence showed that emerging SARS-CoV-2 variants resist neutralizing activity induced by the original SARS-CoV-2 strain.33 Non-RBD binding antibodies induced after a single dose of the BNT162b2 vaccine were reported to be unable to mediate neutralizing activity but could mediate ADCC.34 Another study also showed that anti-RBD antibodies correlated with potent neutralization and weak ADCC activity, whereas antibodies elicited by the S2 subunit correlated with weaker neutralization and stronger ADCC activity.27 This suggests that non-NAbs targeting the S2 domain might have a potential ADCC activity. Therefore, Fc-dependent ADCC effector functions might help identify antigenic targets with extra-neutralizing Fc-mediated activity, as we have shown that multiple antigens can mediate ADCC responses to SARS-CoV-2 with high activity levels. The underlying mechanisms behind such observations remain unclear. However, it seems plausible that specific characteristics of the induced antibodies play an important role.
Notably, omicron infection showed a substantially reduced ability to neutralize SARS-CoV-2 and induce ADCC against SARS-CoV-2 proteins, including WT S, omicron S, and N proteins. Nevertheless, mRNA-vaccinated people with omicron BTI showed high neutralization ability, most likely induced by the mRNA vaccine, as shown by previous studies.35-37 In these studies, higher reductions in neutralization were seen in unvaccinated individuals compared with BTIs, suggesting that the preserved activity against VOCs such as omicron is not a result of higher starting titers but that the quality of NAbs obtained from BTIs is intrinsically better.37 This could be due to continuing affinity maturation or expansion of cross-reactive responses following subsequent antigen exposure.38 Further, we identified that infection with the omicron variant shows significant ADCC escape. This suggests that similar to neutralization, the sequence change in the S protein also affects the quality of antibodies mediating Fc effector function.39, 40 Although ADCC and other Fc effector functions have shown notable resilience despite spike mutations,39 the fact the omicron showed notably diminished susceptibility to ADCC responses implies constraints on this tolerance and offers intriguing perspectives on the specific antibody targets involved. Further, this decreased sensitivity to ADCC indicates that the mutated regions within this variant could be critical in defining significant ADCC epitopes, potentially distinct from those targeted by neutralization. Hence, identifying these specific sites will be crucial in understanding the focal points of antibodies driving ADCC responses.
Interestingly, we showed that symptomatic and asymptomatic infected patients with WT SARS-CoV-2 induced higher ADCC responses against the N protein than omicron infection and vaccination with inactivated virus-based vaccine. The inactivated virus-based vaccine is a first-generation inactivated whole virion vaccine that can induce immune responses to multiple viral proteins, including the N, S, and membrane (M). However, our data showed that it induced weak humoral responses, including binding antibodies and NAbs, compared to natural infection and mRNA vaccines. Other studies also reported that people receiving vector-based and inactivated virus-based vaccines elicit mild antibody responses compared to mRNA vaccines.41, 42 Further, we observed that S-specific ADCC responses induced by the inactivated vaccine were comparable to the ones generated by the other three vaccines but significantly less than those generated by WT SARS-CoV-2 infection.
It is not surprising that S-specific antibodies can stimulate ADCC responses, given that the S protein mediates virus entry into host cells, and this observation is consistent with previous findings.27, 32 However, the N protein is located in virions and is abundantly expressed in infected cells during the early phase of infection; hence, it is difficult for anti-N antibodies to identify infected cells containing the N protein and induce ADCC responses.32 Recovered COVID-19 patients were shown to have high titers of N-specific antibodies.43 However, the functional relevance is still controversial, as there is a lack of knowledge regarding host interactions with the N protein. Anti-N antibodies may only represent passive immune reactions rather than active immune responses to combat SARS-CoV-2 infection, similar to influenza A, where the role of anti-N antibodies was debatable.44 In vitro neutralization studies that attempted to understand the mechanism of anti-N antibody-mediated protection were futile, as these antibodies were consistently found to be non-neutralizing.45, 46 Remarkably, our data clearly showed anti-N antibodies to have the potential to stimulate ADCC responses following SARS-CoV-2 infection, which remained detectable for several months in symptomatic and asymptomatic patients. A recent study reported that N-specific antibodies inhibit complement hyperactivation, improving disease outcomes.47 In contrast, anti-N antibodies might also be involved in a more severe disease progression due to antibody-dependent enhancement.48 Due to the high homology between the N protein of SARS-CoV-2 and other highly pathogenic coronaviruses, Previous exposure could lead to the production of non-neutralizing Abs and establish an overwhelming proinflammatory state through ADE.49 This study showed that N-specific antibodies can activate NK cells and induce ADCC. Whether these responses play a role in protecting against SARS-CoV-2 infection needs further investigation.
The capacity of antibodies to induce Fc-effector functions depends upon antibody specificity, isotype, subtype, and glycosylation.26 Hence, Fc-mediated activities cannot be predicted based on binding and NAbs. Animal models of SARS-CoV-2 infection have shown that the efficacy of monoclonal antibody therapy in mice and hamsters depended on the induction of Fc-dependent effector functions.50, 51 However, the inherent neutralization potency and binding antibody titers might also affect the ability to induce these effector functions. A recent study suggested that the requirement of Fc-dependent effector functions is often reduced in the presence of effective virus neutralization.52 This could explain the weak correlation we obtained between NAbs and ADCC activity targeting the SARS-CoV-2 S protein. Furthermore, a recent study reported that Fc-mediated ADCC responses were strongly correlated with FcγR-binding antibodies, including IgG1 and IgG3.53 It is worth mentioning that antibody fucosylation might also play a role in this regard, leading to differences between infection-induced and vaccine-induced antibodies mediating ADCC responses. Afucosylated IgG binds to its FcγRIII receptor more efficiently. Given that these receptors are expressed on myeloid and NK cells, this leads to enhanced cytokine production and induction of cellular responses, including ADCP and ADCC, upon binding.54 A study assessing ADCC function also showed that peak ADCC levels were correlated with the highest levels of afucosylation in COVID-19 patients.55 This suggests that the enhanced functionality of afucosylated antibodies could drive ADCC responses. Another study assessing afucosylated anti-S IgG1 response induced by the BNT162b2 mRNA vaccine reported that the vaccine induces a transient afucosylated response which decreased within 4 weeks to the level of total IgG1.55 This afucosylated response was similar but less pronounced than observed in natural SARS-CoV-2 infections. Thus, this could explain the differences in ADCC levels seen between natural infection and mRNA vaccination.
In conclusion, our study demonstrates that natural WT SARS-CoV-2 infection and vaccination elicit antibodies capable of inducing NK-cell–mediated ADCC activity. However, the antibodies elicited by WT SARS-CoV-2 are more effective in triggering ADCC than vaccination. These findings could explain the therapeutic effects of plasma with low neutralizing capacity. Also, the fact that mRNA vaccination elicits a higher antibody response, including neutralizing response, but less ADCC activity than natural infection requires further investigation. Additionally, the decreased susceptibility of the omicron variant to ADCC, in contrast to WT-SARS-CoV-2, offers valuable guidance for forthcoming efforts to identify the specific targets of antibodies facilitating ADCC. Last, it is essential to highlight that while ADCC effectiveness against omicron was diminished, any remaining activity might still play a role in protection from severe disease.
AUTHOR CONTRIBUTIONS
The study was conceptualized by Hadeel T. Zedan, Hadi M. Yassine, and Gheyath K. Nasrallah. Duaa W. Al-Sadeq, Eleonora Nicolai, Ali Ait Hssain, Sara Taleb, Hamda Qotba, Khodr Issa, and Laith J. Abu Raddad coordinated sample collection. Experiments were carried out by Hadeel T. Zedan, and Maria K. Smatti. Data processing and statistical analyses were done by Hadeel T. Zedan. Hadeel T. Zedan, Maria K. Smatti, and Hadi M. Yassine did data interpretation and drafting of the manuscript. Gheyath K. Nasrallah, Hadi M. Yassine, and Asmaa A. Althani provided funding. All authors revised and approved the submitted version.
ACKNOWLEDGMENTS
We thank the virology research center at the National Institute of Health (VRC-NIH) for providing the plasmids used to generate SARS-CoV-2 pseudoviruses and for providing HEK293T cells. We also thank the UREP students who assisted in the blood samples collection and coordination: Tala Jamaleddin, Huda Abdul Hameed, Amira Elsharafi, Fatima AlHamaydeh, Bushra Abu Halawa, Hadiya Khalid, Nasrin Cusman, Maram Ali, Hamas Fouda, Salma Mohamud, and Reham Kamal. This study was supported by partial funds from grant #NPRP11S-1212-170092 awarded to Dr. Yassine. GKN would like to acknowledge that this work was made possible by WHO grant number COVID-19-22-43; QUCG-BRC-23/24-170, and grant number UREP29-026-3-004 from the Qatar National Research Fund (a member of Qatar Foundation). The statements made herein are solely the authors' responsibility. Qatar University Open Access publishing facilitated by the Qatar National Library, as part of the Wiley - Qatar National Library agreement
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions. The data supporting this study's findings are available from the corresponding author upon reasonable request.




