Neutralizing antibody response to SARS-CoV-2 bivalent mRNA vaccine in SIV-infected rhesus macaques: Enhanced immunity to XBB subvariants by two-dose vaccination
Abstract
The evolution of SARS-CoV-2 paired with immune imprinting by prototype messenger RNA (mRNA) vaccine has challenged the current vaccination efficacy against newly emerged Omicron subvariants. In our study, we investigated a cohort of macaques infected by SIV and vaccinated with two doses of bivalent Pfizer mRNA vaccine containing wildtype and BA.5 spikes. Using a pseudotyped lentivirus neutralization assay, we determined neutralizing antibody (nAb) titers against new XBB variants, i.e., XBB.1.5, XBB.1.16, and XBB.2.3, alongside D614G and BA.4/5. We found that compared to humans vaccinated with three doses of monovalent mRNA vaccine plus a bivalent booster, the monkeys vaccinated with two doses of bivalent mRNA vaccines exhibited relatively increased titers against XBB subvariants. Of note, SIV-positive dam macaques had reduced nAb titers relative to SIV-negative dams. Additionally, SIV positive dams that received antiretroviral therapy had lower nAb titers than untreated dams. Our study underscores the importance of reformulating the COVID-19 vaccine to better protect against newly emerged XBB subvariants as well as the need for further investigation of vaccine efficacy in individuals living with HIV-1.
1 INTRODUCTION
The Omicron sublineage of SARS-CoV-2 has displayed increasing extents of escape of neutralizing antibodies (nAb) stimulated by messenger RNA (mRNA) vaccination that persists even with new bivalent formulations.1-7 Administration of the three-dose mRNA vaccine can cause immune imprinting, resulting in reduced nAb titers against Omicron subvariants.8, 9 Bivalent vaccination can alleviate this reduction but only to a limited extent, especially against the newer XBB-lineage subvariants.8-10
The disease burden of COVID-19 has affected some populations more than others. One group are individuals living with HIV-1.11 People with advanced and/or untreated HIV-1 infection do not mount as effective as an immune response upon mRNA vaccination compared to treated counterparts.12 Fortunately, individuals receiving antiretroviral therapies (ART) do not appear to be at as high a risk for severe disease.13, 14 Currently, knowledge related to efficacy of mRNA vaccination against SARS-CoV-2 in individuals living with HIV-1 remains limited.
2 RESULTS
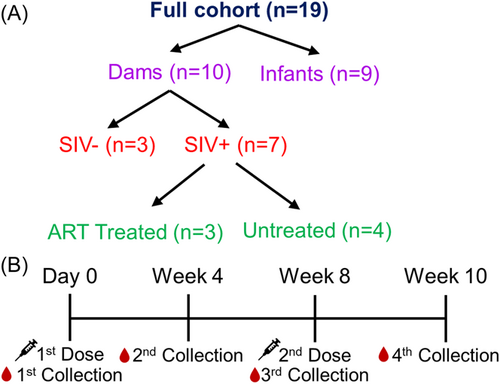
In this study, we investigated the nAb response in a cohort of rhesus macaques including 10 dams (n = 10) and 9 infants (n = 9) (Figure 1A, Tables 1 and 2). Among the dams, seven were intravenously infected with SIV during the first trimester. Of these 7, three received ART during the early third trimester while the remaining four were untreated; ART was interrupted for over 1 year during the priming with the COVID vaccine (Table 2). Notably, none of the infants born to SIV-infected dams tested positive for SIV (Table 2). All monkeys received two shots of Pfizer bivalent mRNA vaccine, which includes both wildtype (WT) and BA.4/5 spikes. An initial dose was administered (day 0) and a booster dose was administered 8 weeks after. Sera were collected at weeks 4, 8, and 10 after the first dose (Figure 1B). Sera were used in a pseudotyped lentivirus neutralization assay15 to determine the nAb response against XBB.1.5, XBB.1.16, and XBB.2.3 variant spikes alongside parental D614G and BA.4/5.
Similar to humans,16 we detected nAb titers against D614G and BA.4/5 at week 4 after the first bivalent dose; titers declined at 8 weeks, but increased after boosting at week 10 (Figure 2A). Titers against XBB variants were near the limit of detection (NT50 = 40) throughout 8 weeks after the first dose, but their levels dramatically increased after boosting, with D614G and BA.4/5 showing a 17–45 and 96–112-fold increase, respectively at week 10 (Figure 2A). The higher titer for BA.4/5 compared to D614G was in sharp contrast to what is seen in humans, where, upon receiving 3 doses of monovalent vaccine and 1 dose of bivalent containing BA.4/5 spike, BA.4/5 titers are usually notably lower than D614G and titers against XBB variants often near-undetectable.1, 4-7 These findings highlight that administration of bivalent vaccine alone can generate robust nAb titers against Omicron subvariants.
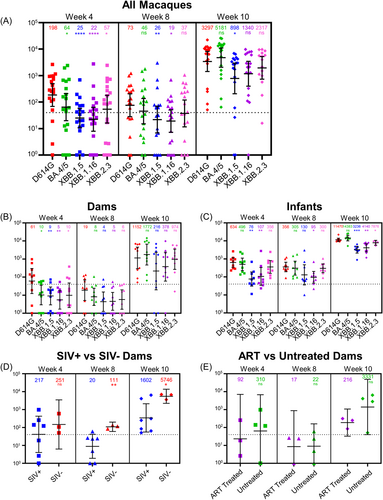
In this cohort, all dams were 8 years of age while infants ranged from 23 to 25 weeks (median 24, Table 1). Similar to humans,17 the nAb response differed dramatically between dams (Figure 2B) and infants (Figure 2C). In dams, the booster dose was required for nearly all monkeys to generate nAb titers above the limit of detection (Figure 2B), while the infants had 5–10-fold higher titers at weeks 4 and 8 against D614G, BA.4/5 and XBB variants, remaining remarkably high at week 10 (Figure 2C). Of note, titers against XBB variants in infants were 22–42-fold higher than that of dams at week 10 (p < 0.0001) (Figure 2B,C), underscoring age as a critical factor for nAb induction during bivalent vaccination.
| Dams (n = 10) | Infants (n = 9) | ||
|---|---|---|---|
| Age (range, median) | |||
| 8 years old | 23–25 weeks (24 weeks) | ||
| SIV exposure | |||
| SIV naive | 3 | SIV naive | 2 |
| SIV positive | 7 | SIV exposed (Born from SIV+ dam, SIV−) | 7 |
| ART treatment | |||
| ART treated | 3 | Born from ART treated, SIV+ dam | 2 |
| Untreated | 4 | Born from untreated SIV+ dam | 5 |
- Note: Summary of Rhesus Macaque Cohort. Details of the macaque cohort are described in the table. SIV naïve dams were never exposed to SIV, and SIV naïve infants were born to uninfected dam cohorts. Infants born to SIV-infected dams were tested as SIV negative.
- Abbreviation: ART, antiretroviral therapies.
Of the total population of dams (n = 10), 7 were SIV positive (Figure 1A and Table 1). For these panels, we focused on nAb titers against D614G, as this variant was included in the bivalent formulation of the vaccine and has been serving as a background/WT for nearly all circulating variants. SIV positive dams had an overall 3.5–5.5-fold lower nAb titer against D614G at 8 and 10 weeks (p < 0.01 and p < 0.05, respectively) relative to uninfected dams (Figure 2D). This aligns with a study conducted in humans demonstrating lower seroconversion in people living with HIV relative to uninfected peers.18 Note that several points for this analysis fell under the limit of detection for the assay (1:40), requiring the extrapolation of these points based on the fit to the curve; thus, statistical significance for the weeks 4 and 8 data should, therefore, be considered with this caveat in mind. Additionally, we found that ART-treated dams had lower titers against D614G at week 10 compared to their untreated counterparts, with a 15.4-fold reduction in titer (p > 0.05) (Figure 2E). This contradicts results seen in humans,12 but could be explained by the interruption of ART therapy and the subsequent resurgence of SIV titers (Table 2) impacting the immune response.
| Infant groups | |||||
|---|---|---|---|---|---|
| Blood (prevaccination) | D614G titer | ||||
| ANML ID | Dam background | PVL (copies/mL) in infant after birth | CD20% | CD4% | Week 10 |
| PN75 | SIV naïve | NA | 7.95 | 60.5 | 12165.4501 |
| PI80 | SIV naïve | NA | 8.3 | 58.2 | 12367.1 |
| PJ44 | SIV infected/untreated | Undetectable | 11.6 | 57.7 | 10526.3158 |
| PJ45 | SIV infected/untreated | Undetectable | 32 | 72.2 | 10689.5 |
| PJ46 | SIV infected/untreated | Undetectable | 26.7 | 63.1 | 9548.05856 |
| PJ28 | SIV infected/untreated | Undetectable | 11.3 | 73.6 | 22209.0613 |
| PH09 | SIV infected/untreated | Undetectable | 11.7 | 59.2 | 9132.4 |
| PI76 | SIV infected/ART treated | Undetectable | 26.5 | 69 | 9230.5 |
| PG73 | SIV infected/ART treated | Undetectable | 28 | 63.1 | 11353.3 |
| Dam groups | |||||
|---|---|---|---|---|---|
| Blood (prevaccination) | D614G titer | ||||
| ANML ID | SIV status during pregnancy | PVL (copies/mL) in adult after C-section and priming | CD20% | CD4% | Week 10 |
| JH11 | SIV naïve | NA | 15.9 | 50 | 6675.6 |
| MI19 | SIV naïve | NA | 12.9 | 47.9 | 3612.7 |
| LR85 | SIV naïve | NA | 15.5 | 66.8 | 6949.3 |
| JN85 | SIV infected/untreated | 8.70E+03 | 46.1 | 41.9 | 6369.4 |
| KJ37 | SIV infected/untreated | 1.40E+05 | 39.4 | 25.5 | 50.9 |
| LM64 | SIV infected/untreated | 2.90E+05 | 45.1 | 35.4 | 4085 |
| LP26 | SIV infected/untreated | 3.50E+04 | 42.8 | 50.8 | 2817.7 |
| JH62 | SIV infected/ART treated | 2.95E+03 | 34.4 | 28 | 84.2 |
| KN45 | SIV infected/ART treated | 4.67E+03 | 61 | 25.6 | 250.2 |
| KL07 | SIV infected/ART treated | 3.62E+03 | 23.3 | 16.2 | 312.7 |
- Note: These tables provide details for the dam and infant portions of the macaque cohort, including the animal identification number (“ANML ID”), color coded by SIV status (green = naïve; orange = SIV infected without ART; blue = SIV infected with ART), plasma viral load (PVL) at time of the first vaccine dose, the percentage of CD20+ B cells (gated lymphocytes) and CD4+ T-cells (gated CD3+ T-cells) in peripheral blood samples using flow cytometry at time of priming, as well as the neutralizing antibody titers against D614G pseudovirus in the week 10 blood collection sample.
3 DISCUSSION
Overall, our findings emphasize a need for further study into the nAb response in people living with HIV to ensure that they remain protected during the ongoing COVID-19 pandemic. The fact that administration of two bivalent mRNA vaccine doses without any monovalent doses yields robust neutralizing antibody titers against D614G and BA.4/5 and strong titers against XBB variants, in contrast to humans that have received 3 doses of monovalent vaccine and a dose of bivalent vaccine,1-7 highlights the potential issue of "immune imprinting" observed in humans as well as the constant need for reformulation of COVID-19 vaccines to improve responses toward future SARS-CoV-2 variants. It is important to note that the monkeys treated with ART had experienced a resurgence in SIV viral load upon pausing of the therapy for vaccine administration, which can put more of a burden on the immune system, thus impacting the ability to mount a response against the SARS-CoV-2 vaccine. This scenario could reflect on individuals living with HIV but inconsistently adhering to ART treatment, a situation that may become more likely upon being infected by SARS-CoV-2. Our study therefore highlights the importance in the continued study of how SARS-CoV-2 and mRNA vaccination impact vulnerable populations including AIDS patients.
4 MATERIALS AND METHODS
4.1 Rhesus macaque cohort
The cohort of rhesus macaques totaled 19 in this study, including 10 dams and 9 infants (Table 1). Seven dams were intravenously infected with SIV at the end of first trimester (~55 days of gestation, dGA), three of which received ART (FTC/TFV/DTG) initiated in the early third trimester (~110dGA) during pregnancy, while 4 remained untreated. The other three dams were SIV naïve throughout the study. Infants were delivered by C-section at 155dGA and all infants born to dam cohorts are tested as SIV negative. The bivalent formulation of SARS-CoV-2 Pfizer mRNA vaccine was administered to all 19 monkeys, including infants (n = 9, 24 weeks of age, SIV negative, Table 2, upper panel), SIV uninoculated adult individuals (n = 3, SIV naïve, ~54.9% of CD4+ T-cell in blood), SIV infected/treated (n = 3, ~23.3% CD4+ T-cells) or untreated (n = 4, ~38.4% CD4+ T-cells) adult animals (Table 2, lower panel). Both the treated and untreated dams showed detectable plasma viral load and chronic status within 13–26 months after C-section. The initial mRNA bivalent vaccine dose (0.3 mL/30 µg) was followed by a booster dose administered 8 weeks later. Blood was collected on the day of vaccination (day 0), 4 weeks after the first dose, 8 weeks after the first dose (at the time of the booster), and 10 weeks after the first dose (2 weeks post-booster). ART was discontinued after C-section, leading to rapid viral rebound in these SIV-infected dams after analytical treatment interruption, establishing a chronic SIV infection status during vaccination.
4.2 Cell lines and maintenance
Cell lines used in this study included HEK293T (ATCC CRL-11268, RRID: CVCL_1926) and HEK293T expressing human ACE2 (HEK293T-ACE2) (BEI NR-52511, RRID: CVCL_A7UK). Cells were maintained in DMEM (Gibco, 11965-092) supplemented with 10% fetal bovine serum (Sigma, F1051) and 0.5% penicillin-streptomycin (HyClone, SV30010). For splitting, cells were washed with phosphate-buffered saline (Sigma, D5652-10X1L) and incubated in 0.05% trypsin + 0.53 mM ethylenediaminetetraacetic acid (Corning, 25-052-CI) until completely detached. Cells were maintained at 37°C and 5.0% CO2.
4.3 Plasmids
Plasmids used in this study include each of the spike expression constructs and an HIV-1 pseudotyping vector. The spike plasmids were engineered in the pcDNA3.1 plasmid backbone and include FLAG tags on the N- and C-termini as well as BamHI and KpnI sites at the 5′ and 3′ ends of the spike insert respectively. D614G and BA.4/5 constructs were cloned by GenScript using restriction enzyme cloning. XBB.1.5, XBB.1.16, and XBB.2.3 were generated in the lab through site-directed mutagenesis via PCR. The HIV-1 vector used in this study is a pNL4-3 strain vector with an Env deletion and secreted Gaussia luciferase reporter gene interrupted by an antisense intron (inGluc).
4.4 Pseudotyped lentivirus production and infectivity
To produce pseudotyped lentivirus, HEK293T cells were cotransfected using a polyethyleneimine transfection (Transporter 5 Transfection Reagent, Polysciences) with inGluc vector and spike plasmid in a 2:1 ratio. Virus was collected by taking media off the transfected cells and storing at −80°C 48 and 72 h posttransfection. Infectivity of the pseudotyped lentiviruses was determined by infecting HEK293T-ACE2 cells with 100 µL of each virus. Luminescence by secreted Gaussia luciferase was used to determine relative infectivity 48 and 72 h postinfection. To measure luminescence, 20 µL of infected cell media was combined with 20 µL luciferase substrate (0.1 M Tris pH 7.4, 0.3 M sodium ascorbate, 10 µM coelenterazine) and read on a BioTek Cytation plate reader. Infectivity readout at 48 h postinfection was used to normalize viral titer for the neutralization assay.
4.5 Virus neutralization assay
Sera taken from the cohort of monkeys was diluted at an initial diluton of 1:40 and serially diluted fourfold (final dilutions 1:40, 1:160, 1:640, 1:2560, 1:10 240, and no serum as a control). Pseudotyped lentivirus was diluted in DMEM + 10% FBS + 0.5% Pen/Strep as described above. A total of 100 µL of normalized virus was added onto the diluted serum samples and incubated for 1 h at 37°C. The virus/sera mixture was then used to infect HEK293T-ACE2 cells. Luminescence was measured as described above at 48 and 72 h postinfection. Neutralization titers at 50% (NT50) were determined using least-squares fit non-linear regression with a normalized response (no serum control). GraphPad Prism v9 was used for these calculations (San Diego, CA).
4.6 Quantification and statistical analysis
All statistical analyses were conducted using GraphPad Prism v9 (San Diego, CA). Of note, 4 infant samples do not have data for XBB.2.3 titers at week 10 which resulted in slightly different analyses for cohort subsets that included these samples. Throughout, significance was determined using log10 transformed NT50 values to better approximate normality. In Figure 2A and 2C, panels (2A and 2C) were separated by timepoint and analyzed using a repeated measures one-way analysis of variance (ANOVA) with Bonferroni posttest except for the week 10 timepoint which was analyzed using a mixed-effects model due to the missing data points. In Figure 2B, panel (2B) was analyzed using the repeated measures one-way ANOVA with Bonferroni posttest. In Figure (2D,E), data were separated by timepoint and a two tailed t-test with Welch's correction was run to determine significance. Significance is displayed as nsp > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
AUTHOR CONTRIBUTIONS
Shan-Lu Liu and Huanbin Xu conceived and directed the project. Julia N. Faraone performed most of the experiments. Xiaolwei Wang, Panke Qu and Eunice Vincent assisted in experiments. Huanbin Xu collected and provided sera samples. Julia N. Faraone, Huanbin Xu, and Shan-Lu Liu wrote the paper. Yi-Min Zheng provided insightful discussion and revision of the manuscript
ACKNOWLEDGMENTS
We thank the NIH AIDS Reagent Program and BEI Resources for supplying reagents that made this work possible. We also thank the members of Liu lab for sharing reagents and helpful discussion. This work was supported by a fund provided by an anonymous private donor to OSU; additional support of S.-L.L.'s lab includes National Cancer Institute U54CA260582. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. H.X. was supported by National Institutes of Health grants R01 AI147372, R01 HD099857, R01 MH133474, and the Office of Research Infrastructure Programs (ORIP) grant no. OD011104.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
ETHICS STATEMENT
All animals in this study were housed at the Tulane National Primate Research Center in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care International standards. All studies were reviewed and approved by the Tulane University Institutional Animal Care and Use Committee under protocol number P0408. Animal housing and studies were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (NIH, AAALAC #000594) and with the recommendations of the Weatherall report: The Use of Non-Human Primates in Research. All clinical procedures were carried out under the direction of a laboratory animal veterinarian. All procedures were performed under anesthesia using ketamine, and all efforts were made to minimize stress, improve housing conditions, and to provide enrichment opportunities (e.g., objects to manipulate in cage, varied food supplements, foraging and task-oriented feeding methods, interaction with caregivers and research staff).
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in NCI SeroNet at https://www.cancer.gov/research/key-initiatives/covid-19/serological-sciences-network.




