The expression analysis of human endogenous retrovirus-K Env, Np9, and Rec transcripts in cervical cancer
Abstract
While infection with high-risk human papillomavirus (HPV) types is necessary for cervical cancer (CC) development, it is not enough, and other risk factors are required. Several studies have reported the activation of HERV-K in different cancers; however, the investigation of HERV-K expression levels in CC is scarce. In this study, it was hypothesized that activation of HERV-K could play an essential role in CC development. In this order, the expression levels of HERV-K Env, Np9, and Rec transcripts were investigated on 147 normal to CC uterine cervical tissues using quantitative real-time PCR. The significantly higher levels of HERV-K Env and Np9 transcripts were found in patients with cervical intraepithelial neoplasia (CIN) II−III and CC groups compared to those in the normal/CIN I group. Expression of Rec transcript was also higher only in the CC group than normal/CIN I group. Among CC patients, meaningfully higher levels of HERV-K Env and Np9 transcripts were found in patients with squamous cell carcinoma rather than in adenocarcinoma. When only the HPV 16 positive samples were investigated, it was found that the mean difference in Env and Np9 mRNA levels was meaningfully higher among precancer lesions and the cancer group in comparison with the normal group. However, the Rec mRNA level showed no significant differences. The association between the expression of HERV-K genes was investigated, and a significant positive correlation of Env expression with Np9 transcript was found only in the group with precancer lesions (R = 0.6, p = 0.0037). Moreover, a significant positive correlation was found between Rec and Np9 transcripts in patients with normal cervix tissues (R = 0.26, p = 0.033). However, no correlations were observed between the expression of Env and Rec in the three groups. In conclusion, our results showed that HERV-K transcripts, especially Env and Np9, upregulated during cervical lesion progression. These findings highlight the potential use of HERV-K Env and Np9 as biomarkers for CC diagnosis and prognosis. Further investigation is needed to determine the clinical utility of these markers and whether targeting HERV-K oncogenes could be a viable therapeutic strategy for CC.
1 INTRODUCTION
Cervical cancer (CC) is defined as the fourth commonly occurring cancer and also the fourth leading cause of cancer death in women. Globally, in 2020, it was estimated that there were 604 127 new cases of women who suffered from CC and about 342 000 women's deaths.1 Worldwide, it was ranked as the second most common cause of cancer incidence and mortality in women of reproductive age.2 In total, about 84% of the CC incidence and 88% of deaths from CC occurs in countries with the lowest values on the Human Development Index.3, 4
It is well-documented that the natural history of the CC carcinogenesis model is made of multistep processes.5 The first stage involves infection by high-risk human papillomavirus (HPV) types, particularly HPV 16 and 18. However, a large proportion of these infections clear within a maximum of 2 years.1 Only a tiny proportion of high-risk HPV infections persist and progress toward cervical intraepithelial neoplasia I−III (CIN I−III) and trigger the next stage of carcinogenesis. While most of the CIN I and II lesions regress and revert to normal cells, in some cases, precancerous lesions progress to CIN III and invasive CC.2
Although infection with high-risk HPV types is necessary for CC development, it is insufficient, and other risk factors are needed.5 It is known that several factors, including early age of sexual activity initiation, the number of sexual partners, smoking, use of oral contraceptive pills, multiple births, history of sexually transmitted infections (particularly human immunodeficiency virus [HIV] and Chlamydia trachomatis), and vaginal microbiota can play an essential role as risk-factor in CC development.6-8 Also, the activation of Human Endogenous Retroviruses (HERV) may be the case.9
HERVs are ancestral retroviral sequences that endogenized into the early primate species germline and integrated into DNA from millions of years ago.10 HERVs currently comprise almost 8% of human chromosomal DNA.11 Though HERVs play a critical role in embryogenesis and the formation of trophoblasts, they are silenced in most cell types in healthy individuals.12 HERVs could be reactivated in various pathological conditions, including breast, prostate, colorectal, and ovarian cancers, showing the ability of HERVs transcripts and proteins to regulate human gene expression and disrupt normal cellular processes and even, in some cases, act as oncogenes.3, 13-16 In this complex scenario, several HERVs, such as HERV-K (HML-2), have been proposed as endogenous carcinogenic factors in various cancers, spanning the bridge between environmental risk factors, genetic susceptibility, and immune response. Indeed, HERVs can lead to cancer through several mechanisms including, genetic alterations in cancer, HERV LTR as an alternative promoter or enhancer of cellular genes, aberrant signaling, and immune suppression which may lead to cancer formation and spreading.17-19
HERV-K proviruses encode two accessory oncoproteins, Np9 (HERV-K type-II) and Rec (HERV-K type-I), that exert their carcinogenesis effects via interaction with cellular transcriptional factors and tumor suppressor proteins.18 Several studies found that the activation of HERV-K oncoprotein transcription enhanced tumourigenesis of multiple tumor viruses, including hepatitis B and C, human cytomegalovirus, Epstein−Barr virus, Kaposi−Sarcoma herpesvirus, and HIV.10 Interestingly, the higher expression levels of HERVs, including HERV-K and HERV-W, have been observed in COVID-19 patients. It is suggested that they could be used as biomarker to predict COVID-19 severity and disease outcome.20-22
While many reports defined the activation of HERV-K in various cancers, research on the investigation of the HERV-K oncoprotein expression level in CC tissues is scarce. On the other side, the identification of various cofactors that accelerate HPV tumorigenesis into precancer lesions and CC will be supportive in the triage of patients in the clinical setting. Given the known immunomodulating and oncogenic features of HERV-K products, we hypothesized that hyperactivation of HERV-K could play an essential role in CC development. If the hypothesis is true, it can be applied as a candidate prognostic or therapeutic biomarker to predict the clinical progression of CC in the future.
2 MATERIALS AND METHODS
2.1 Study population
This cross-sectional, double-center study involved patients from two academic referral gynecology centers in Tehran, Iran, from 2020 to 2022. This study recruited 148 fresh biopsies of the uterine cervix obtained by colposcopy or hysterectomy from women who were visited by two attending physicians in Yas and Imam Khomeini Hospitals. Women who received radiation or chemotherapy were excluded from this study. One patient with HIV infection undergoing HAART therapy was excluded from this study.
The mean age of 147 studied individuals was 39.5 ± 12 years, with a range of 21−86 years. Specimens were stratified into three distinct pathological categories, including: 78 normal or CIN-I (mean age of 35.6 ± 8.4 years; ranging from 21 to 61 years); 29 CIN II−III (mean age of 37.2 ± 10.6 years; ranging from 22 to 66 years); and 40 CC samples (mean age of 49.1 ± 14.1 years; ranging from 30 to 86 years).
Clinical and demographic information was collected through a questionnaire. All the procedures followed the ethical guidelines of the Helsinki Declaration and were approved by the Tehran University of Medical Sciences (TUMS) (IR.TUMS.SPH.REC.1399.235). All subjects signed the informed consent that was approved by the local ethical committee of TUMS.
2.2 Tissue RNA/DNA extraction and cDNA synthesis
All tissue biopsies were merged into RNA Later (GeneAll Co.), stored overnight at +4°C, and then transferred to −80°C until nucleic acid extraction. RNA/DNA extraction was performed by TRIZOL reagent (GeneAll Co.) as described by the manufacturer's instruction. At the final elution step, extracted RNA and DNA were eluted with 40 μL DNase/RNase-Free DEPC-treated water, and the aliquots were stored at −80°C and −20°C, respectively. The DNA and RNA purity and concentration were determined using the NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific). To eliminate contaminating genomic DNA, extracted RNAs were treated with RNase-free DNase I (Thermo Fisher Scientific) according to manufacturer's instructions. Total RNAs of all samples were then adjusted to the same final concentration, and 1 µg of RNAs were reverse-transcribed using the SinaClon First Strand cDNA Synthesis Kit (Sinaclon Co.), according to the manufacturer's instructions.
2.3 HPV DNA detection and genotyping
All samples were screened for HPV by nested polymerase chain reaction (nested-PCR) using primer pairs of MY09/11 and GP+5/+6, as described previously.23 Briefly, 200 ng of DNA template was subjected to the amplification of a 150 bp fragment of the L1-coding gene in a reaction mixture containing 10 pmol of each primer pair, 50 μM of each dNTP, 2 mM MgCl2, and 1 U of Taq-DNA polymerase. Thermal cycles were as follows: 35 amplification cycles at 95°C for 30 s, 55°C (first round) and 50°C (second round) for 45 s, and 72°C for 1 min. Negative control (sterile water) was included in every set of PCR runs. The PCR products were loaded on the 2% agarose gel. All HPV-positive samples were subjected to bidirectional direct sequencing via a BigDye® Terminator v3.1 Sequencing Kit and a 3130 Genetic Analyzer Automated Sequencer (Applied Biosystems) according to manufacturer's instructions. Nucleotide sequences were blasted using the NCBI Blast server to distinguish HPV genotypes.
2.4 Relative quantification by real-time PCR
Target genes (Envelope [Env], Rec, and Np9)24 were relatively quantified by Real-Time PCR using SYBR® Premix Ex Taq™ master mix (Takara Bio) on the StepOne Plus Real-Time PCR system (Applied Biosystems). Briefly, PCR amplification reactions were performed in 20 µL reaction mixtures containing cDNA (10-fold diluted), 2x SYBR Green master mix, 50× ROX dye, and 10 pmol of each primer pair (Table 1). The reactions were incubated at 95°C for 2 min, followed by 40 cycles of 95°C for 15 s, 57°C for 30 s, and 72°C for 10 s. Distilled water25 was used as a negative control. A melting curve analysis was performed to confirm single gene-specific peaks by heating samples from 72°C to 98°C at the end of the amplification cycles. The linearity and accuracy of Real-Time RT-PCR were assessed using the GAPDH (housekeeping gene) standard curve derived from amplifying serially diluted pooled cDNA (a 10-fold dilution series). Expression of Env, NP9, and Rec mRNAs was investigated according to the method, and the levels of each gene were normalized to the GAPDH cDNA amount. The relative value of each target gene was expressed as the fold-change of the case group over the control group to compare mRNA levels among the groups.
| Primer | Strand | Start | Stop | Sequence | Product length | Reference |
|---|---|---|---|---|---|---|
| HERV-K Env | 5′-Forward | 6870 | 6887 | CTGCCCTGCCAAACCTGA | 186 | |
| 3′-Reverse | 7055 | 7038 | AGTGACATCCCGCTTACC | [24] | ||
| HERV-K Np9 | 5′-Forward | 8404 | 8426 | AGATGTCTGCAGGTGTACCCA | 119 | [24] |
| 3′-Reverse | 8522 | 8501 | CTCTTGCTTTTCCCCACATTTC | |||
| HERV-K Rec | 5′-Forward | 6506 | 6529 | ATCGAGCACCGTTGACTCACAAGA | 172 | |
| 3′-Reverse | 6657 | 6677 | ATACTCTCTGGGGTTTGTGTC | [24] | ||
| GAPDH | 5′-Forward | 7029 | 7048 | ACCAGGGCTGCTTTTAACTC | 151 | |
| 3′-Reverse | 7179 | 7158 | TGACAAGCTTCCCGTTCTCAG | - |
- Abbreviations: GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HERV, human endogenous retrovirus.
2.5 Statistical analysis
The statistical analysis was performed with GraphPad Prism 9.0 (GraphPad Software). Normally distributed numeric variables were assessed via two-tailed Student's t-test and Welch ANOVA test. Non-normally distributed variables were calculated by Mann−Witney t-test and Kruskal–Wallis ANOVA post hoc variance analysis. Receiver operating characteristic (ROC) and area under the ROC curve (AUC) were prepared and analyzed to estimate the diagnostic value using R software (R- Version 4.3.1). Statistical significance was set at *p < 0.05, **p < 0.01, and ***p < 0.001.
3 RESULTS
3.1 Demographic characteristics and HPV genotyping
In this study, 147 uterine cervix biopsies were investigated, including 78 normal/CIN-I, 29 CIN II−III, and 40 CC samples. Considering the types of CC, squamous cell carcinoma (SCC) and adenocarcinoma (AdCa) were confirmed by the pathological examination in 32 (80%) and 8 (20%) samples, respectively.
HPV was detected in 92.3% of normal/CIN I, 90% of CIN II−III, and 95% of CC samples. The prevalence of HPV types was as follow: in normal/CIN I group, HPV 16 (41%), HPV 18 (12.8%), and other types (38.5%); in CIN II−III group, HPV 16 (56.7%), HPV 18 (13.4%), and other types (19.9%); and in CC group, HPV 16 (60%), HPV 18 (20%), and other types (15%).
3.2 Expression of HERV-K Env, Np9, and Rec mRNAs about histology
The mRNA levels of Env, Np9, and Rec were normalized and analyzed by the comparative CT method. The higher levels of Env and Np9 transcripts were found in the CIN II−III group compared with the normal/CIN I group, and the differences were statistically significant (p = 0.0004 and p = 0.0003, respectively). The expression levels of Env and Np9 were also significantly increased in the CC group when compared with the normal/CIN I group (p = 0.006 and p = 0.0025, respectively) (Figure 1A,B). Although mRNA expression of Rec was higher in the CC group than the normal/CIN I group (p = 0.0335), no statistically significant difference was observed between CIN II−III and normal/CIN I groups (Figure 1C). It is necessary to be mentioned that no or weak amplification of HERV-K Rec mRNA was observed in some cases.
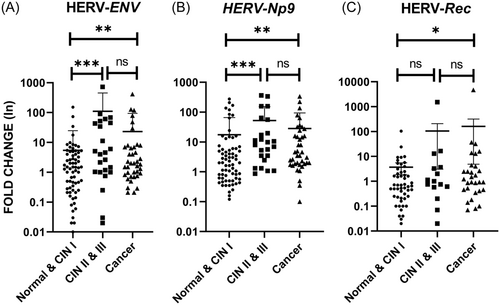
Considering the stratification of CC to SCC and AdCa groups, the higher expression levels of Env and Np9 mRNAs were observed between these two groups as a statistically significant difference was found (p = 0.032 and p = 0.0248, respectively) (Figure 2A,B). However, the expression level of Rec transcript was marginally nonsignificant between SCC and AdCa cases (p = 0.0508) (Figure 2C).

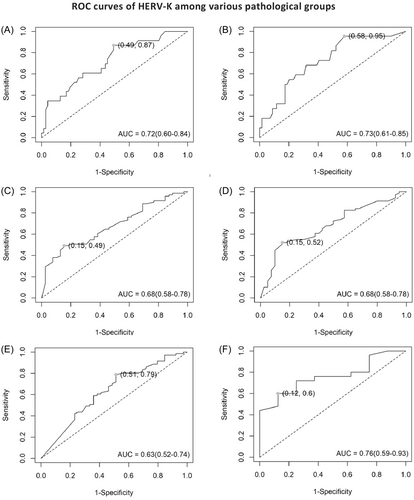
ROC was used to assess the diagnostic accuracy of higher expression of HERV-K Env, Np9, and Rec in precancer and cancer groups in comparison to controls. The AUC revealed an excellent separation between women with normal cervix tissue and precancer lesions, considering the expression of Env (Figure 3A) and Np9 (Figure 3B) mRNAs (Env: AUC = 0.72; 95% confidence interval [CI]: 0.6−0.84 with 87% sensitivity and 50% specificity, p < 0.001; Np9: AUC = 0.73; 95% CI: 0.61−0.85 with 96% sensitivity and 42% specificity, p < 0.001). HERV-K Rec mRNA did not show any significant predictive values between these two groups (data not shown). Between CC and normal groups, the significant diagnostic value was also determined for Env, Np9, and Rec transcripts, which correctly distinguished samples with CC from samples with normal histopathology (Figure 3C−E). The ROC curve AUC of both Env and Np9 was 0.68 (95% CI: 0.58−0.78, p < 0.001 for both Env and Np9). The AUC for Rec was 0.63 (95% CI: 0.52−0.74, p < 0.001). Interestingly, when CC patients stratified into SCC and AdCa groups, only the level of Env (Figure 3F) could be considered as a potentially valuable candidate biomarker in discriminating SCC from AdCa (AUC = 0.76, 95% CI: 0.58−0.78 with 84% specificity and 50% sensitivity, p < 0.001).
3.3 Expression of HERV-K Env, Np9, and Rec mRNAs corresponding to HPV genotypes
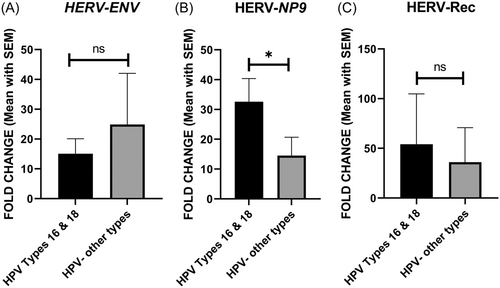
All HPV-positive samples were classified into two groups: HPV 16/18 positive and samples infected with other HPV types. As shown in Figure 4, the expression level of Np9 was significantly higher in HPV 16/18 positive samples compared to the other group (p = 0.0462). However, there was no significant difference in the expression levels of Env and Rec between the two groups (Figure 4).
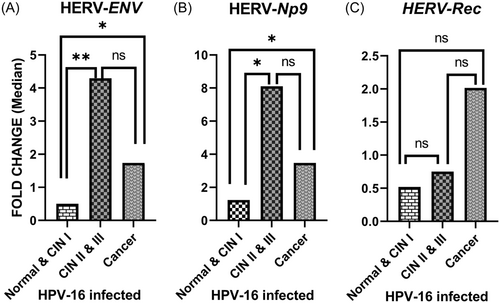
When only the HPV 16 positive samples were stratified into three pathological groups (normal, precancer, and cancer), it was found that the mean difference in Env and Np9 mRNA levels were meaningfully higher among precancer lesions (p = 0.0056 and 0.0067, respectively) and cancer group (p = 0.027 and 0.0208, respectively) in comparison with the normal group (Figure 5A,B). However, the Rec mRNA level showed no significant differences (Figure 5C).
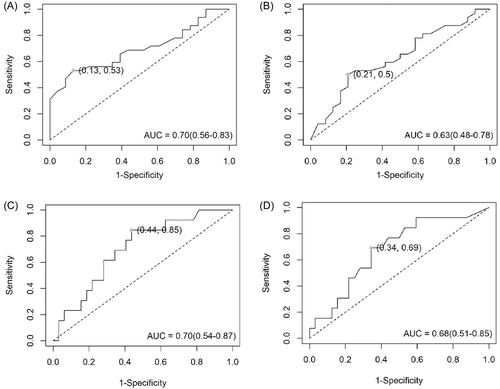
The AUC curve was applied to define the diagnostic accuracy of HERV-K Env, Np9, and Rec mRNAs expression among HPV 16-infected patients. Significant diagnostic value was displayed for Env (Figure 6A) and Np9 (Figure 6B) mRNAs between cancer and normal cervix tissue groups (Env: AUC = 0.7; 95% CI: 0.56–0.83 with 53% sensitivity and 87% specificity, p = 0.001; Np9: AUC = 0.63; 95% CI: 0.48–0.78 with 50% sensitivity and 79% specificity, p = 0.05). However, Rec mRNA expression did not show significant diagnostic values among cancerous and normal cervix groups (data not shown). Consequently, the AUC curve for Env (AUC = 0.7, 95% CI: 0.54–0.87 with 84% specificity and 70% sensitivity, p = 0.018) and Np9 (AUC = 0.68, 95% CI: 0.58–0.78, with 84% specificity and 50% sensitivity, p = 0.032) mRNAs correctly distinguished women with precancer lesions from women with normal cervix tissue (Figure 6C,D).
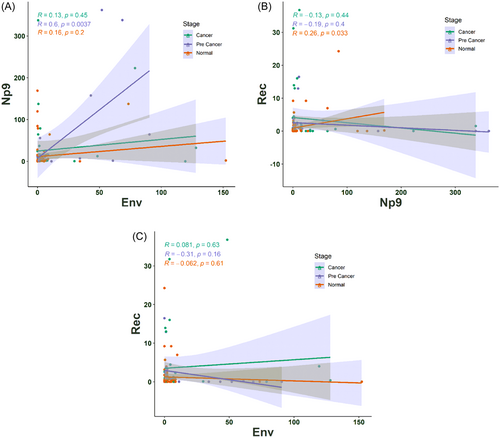
We also examined whether HERV-K gene transcripts are in association with the expression level of each other in a series of patients from normal to CC tissues. The association between the expression of HERV-K genes was investigated, and a significant positive correlation of Env expression with Np9 transcript was found only in the group with precancer lesions (R = 0.6, p = 0.0037; Figure 7A). Moreover, a significant positive correlation was found between Rec and Np9 transcripts in patients with normal cervix tissues (R = 0.26, p = 0.033; Figure 7B). As depicted in Figure 7C, no correlations were observed between expression of Env and Rec in the three groups.
3.4 Expression of HERV-K Env, Np9, and Rec mRNAs regard several variables
The association of HERV-K Env, Np9, and Rec mRNAs expression and clinic-pathologic characteristics of patients are shown in Table 2. The mRNA levels of HERV-K Env and Np9 indicated a statistically significant difference relative to age (p = 0.0302 and 0.0272, respectively) as the higher expression level of these two transcripts was found at age >36 years old. The expression level of Rec transcripts was slightly higher in older patients; however, this difference was not found to be statistically significant (p = 0.223).
| Fold change of HERV-K mRNAs | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Variables | Env | Np9 | Rec | |||||||
| N | Mean ± SD | p | N | Mean ± SD | p | N | Mean ± SD | p | ||
| Age (year) | <35 >36 |
55 75 |
5.8 ± 13.8 27.8 ± 98.7 |
0.0302** | 55 75 |
16.03 ± 39.9 34.9 ± 80.9 |
0.0272** | 55 75 |
3.9 ± 14.2 22.5 ± 176.6 |
0.405 |
| Number of pregnancies | ≥4 ≤3 |
21 46 |
7.3 ± 16 8.03 ± 23.1 |
0.388 | 22 44 |
33.4 ± 80.3 5.8 ± 7.4 |
0.028* | 21 44 |
3.3 ± 8.4 1.7 ± 4.3 |
0.315 |
| Number of births | ≥3 ≤2 |
18 42 |
4.42 ± 11.1 7.3 ± 17.8 |
0.721 | 20 42 |
13.4 ± 34.7 32.2 ± 79.7 |
0.785 | 18 43 |
3.8 ± 9.05 1.4 ± 4.1 |
0.041* |
| Number of abortions | ≥2 ≤1 |
10 21 |
8.7 ± 21.8 8.07 ± 18.8 |
0.843 | 10 22 |
5.4 ± 8.2 46.1 ± 100 |
0.025* | 10 21 |
3.3 ± 6.6 1.06 ± 1.7 |
0.5 |
| Tumor TNM stage | I−II III−IV |
10 4 |
11.8 ± 23.8 1.9 ± 1.6 |
0.239 | 10 4 |
35.8 ± 68.2 4.4 ± 2.8 |
0.227 | 9 3 |
2.6 ± 4.5 10.5 ± 7.04 |
0.031* |
- Abbreviations: HERV, human endogenous retrovirus; SD, standard deviation.
- *p < 0.05; **p < 0.01.
The level of Np9 transcript also significantly increased concerning the number of pregnancies ≥4 (p = 0.028). However, the expression of Np9 was significantly decreased in proportion to the number of abortions ≥2 (p = 0.025). The expression level of Rec transcript showed a dramatic increase as per the number of births ≥3 (p = 0.041) and tumor TNM stage III−IV (p = 0.031).
4 DISCUSSION
The current study investigated the expression levels of three HERV-K (HML-2) transcripts (Env, Np9, and Rec) in uterine cervical biopsies, ranging from normal to CC. The results of this study showed significant upregulation of the three transcripts in cervical precursor lesions and cancer groups in comparison to normal cervical tissues. In agreement with our results, the activation of HERV has been shown in various cancers such as breast,26 ovarian,16 lymphoma,26, 27 melanoma,28 prostate, sarcoma, colon, and leukemia.29-31 It has also been suggested that the expression profile of HERV-K oncogenes can differentiate low-grade from more aggressive cancerous forms, especially in prostate cancer,32 multiple myeloma,33 breast cancer,13 and hepatocellular carcinoma.15 However, it is not yet clear whether activation of HERVs induces cancer development or whether activation of HERV is a bystander effect of altered gene regulation in the cancer microenvironment.18
The higher expression levels of Env and Np9 were also shown among SCC than AdCa patients. The ROC curve findings confirmed the potential diagnostic value of HERV-K transcript expression during CC progression, indicating that HERV-K transcripts could serve as a potential biomarker in the diagnosis and prognosis of CC.
There are potentially complex mechanisms underlying the association between HERVs and cancers. Some of these features include the tissue-specific manner of HERV-K expression,34 promotion of cell growth and proliferation,35 expression of HERV-K gene biomarkers alongside tumor disease progression in several cancers, such as breast cancer,36 multiple myeloma,33 and CC, as well as transactivation of HERV-K due to the presence of tumor-specific transcription factors.16 Indeed, Kaposi's sarcoma-associated herpesvirus (KSHV)-related latent proteins LANA and v-FLIP elicit HERV-K transactivation through interaction with the MAPK-ERK and NF-kB signaling pathways and SP1 transcription factor. This hyperactivation, in turn, leads to the promotion of Kaposi's sarcoma cell growth.24 Another possible route of HERV-K hyperactivation could be due to global hypomethylation in HERV-K DNA during tumor transformation. Accordingly, it has been shown that selective hypomethylation of HERV-K resulted in a quantitatively higher HERV-K expression in ovarian cancer tissues at both transcriptional and translational levels compared to no expression in normal tissues.16
Overall, no relationship between HERV-K expression and different HPV types was observed in this study. However, the levels of Env and Np9, but not Rec, differed significantly among cervical precancer and cancer groups compared to the normal group in HPV 16-infected patients than other HPV types. This finding suggests that HERV-K mRNA expression may interact more specifically with certain HPV genotypes such as HPV 16. The AUC curve among HPV 16-infected patients showed a significant diagnostic value for Env and Np9 mRNAs between cancer and normal cervix tissue groups. Interestingly, a recent study based on RNA-seq data investigated the expression of ERVs in CC. They revealed that ERV expression had the highest correlation in the sample with active HPV compared to other variables such as ethnicity and disease stage.37 It has been demonstrated that a combination of HERV expression signatures with clinical data, such as HPV status, results in a significant improvement of prognostic power compared with the clinical factors alone.37
Almost all CCs involve the high-risk HPV oncoproteins as a distinct virally-mediated carcinogenic mechanisms; nevertheless, the role of HERVs in HPV tumor microenvironment modulation, and especially, the mechanism by which HERVs affect the integration of the HPV genome remains unknown. One hypothesis suggests that HPV integration may awaken HERVs in a way similar to the hyperactivation observed in the triggering of HERV-K with various viral proteins such as KSHV LANA and v-FLIPs.24 One study by Curty et al. investigated the effect of HPV integration on HERV expression in a limited number of samples and found too far genetic distance between HERV loci and the HPV integration site.9 Further transcriptional and translational studies with a large number of patients could greatly clarify this topic, as HPV is capable of integrating randomly, which possibly will affect HERV oncogene expression.
The modulation of immune response in the tumor microenvironment by HERV-K oncogenes may affect HPV persisten, consequently can lead to the development of cancer. HERV-K Env is thought to enable the fusogenic of HPV-infected cancer cells and create immunosuppressive conditions that allow for immune evasion of HPV-infected cells and help tumor expansion.38 Moreover, HERVs provide microRNA responsive elements (MRE), essential loci for the complementary binding, which act as a reservoir of tumor suppressive-related miRNAs and lncRNAs. The lncRNAs play essential roles as either tumor suppressor (let, gas5) or oncogene-related (MALAT1), and their up/downregulation have been demonstrated in CC.39 Furthermore, DNA methylation is an epigenetic mechanism that controls HERV expression. Curty et al. showed a significantly negative correlation between DNA methyltransferase expression and expression of retroelement, such as L1, during CC progression.9 More importantly, Iramaneerat et al. found HERV-K significantly hypo methylated in 29 ovarian clear cell carcinoma patients.40 Likewise, alterations in HERV methylation may occur during CC progression as a result of HR-HPV infections. In particular, both HPV and HERV-K noncoding regions have a distinct element, the progesterone response element, which displays responsiveness to the progesterone hormone lead to gene transactivation.41, 42 Consequently, the occurrence of hypomethylation in these regulatory regions may enhance the oncogenic properties of these viruses in cervix tumorigenesis through the generation of genetic instabilities, promoting cancer stemness, and carcinogenesis. Although, to the best of our knowledge, no study had been conducted in this matter.
In addition, HERV-K expression, among many factors, is associated with cancer cell plasticity and stemness maintenance. HML-2 LTR sequences are seeded with transcription factors binding motifs that contribute to altering neighboring gene expression. An active binding motif for a nuclear transcription and pluripotency factor, octamer-binding transcription factor 4 (OCT4), has been detected on the HERV-K LTR 43 which recruits OCT-4 to further activate HML-2 viral transcripts. The interaction of HERV-K with OCT-4 during carcinogenesis further enhances neighboring gene expression such as pro-growth genes to induce cell proliferation.41 Also, in breast and liver cancer cells, a strong positive correlation of different embryonic genes such as OCT-4 and NANOG homeobox (NANOG) has been demonstrated due to the HERV-K overexpression. Indeed, embryonic genes play a role in acquiring cancer stemness characteristics through the cell proliferation process and initiation of cell migration.44
The most important limitations of this study were the modest sample size and lack of functionally confirmation of our findings. We are suggesting that the expression levels of E6/E7 proteins and integration status of HPV will be investigated among HPV 16 and 18-infected women. Also to clarify whether E6/E7 oncoproteins activate HERV-K or not, suggesting use of siRNAs against these proteins in future studies.
In conclusion, our findings showed the expression of HERV-K mRNAs, especially Env and Np9, was upregulated during the progression of cervical lesions. Also, the levels of Env and Np9, but not Rec, differed meaningfully among cervical precancer and cancer groups in comparison to the normal group in HPV 16-infected patients than other HPV types. HERV-K may play a role in promoting the expression of HPV oncogenes, accelerating HPV integration, modulating the immune response to HPV, promoting cell growth, and poor prognosticating in CCs. Our study provided important insights into the role of HERV-K expression in the severity of cervical precancer and cancer development and highlights the potential use of HERV-K Env and Np9 as biomarkers for CC diagnosis and prognosis. Further investigation is needed to determine the clinical utility of these markers and whether targeting HERV-K oncogenes could be a viable therapeutic strategy for CC.
AUTHOR CONTRIBUTIONS
Arash Arashkia and Somayeh Jalilvand have contributed to the design of the study. Rahim Soleimani-Jelodar and Kimia Sharifian have done all tests. Setareh Akhavan, Fariba Yarandi, and Fatemeh Nili have contributed to the diagnosis and confirmation of pathological results. Mohammad Farahmand has contributed to the analysis and interpretation of data. Somayeh Jalilvand and Zabihollah Shoja have contributed to drafting the article.
ACKNOWLEDGMENTS
This study has been funded and supported by the Tehran University of Medical Sciences (TUMS), Grant no. 99-3-99-50307, and the Iran National Science Foundation (INSF) (Grant no. 99024518). It has also been part of a PhD thesis supported by Tehran University of Medical Sciences; Grant no. 240/752.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ETHICS STATEMENT
Informed consent was obtained from all study subjects and the study was approved by the local ethical committee of Tehran University of Medical Sciences (IR.TUMS.SPH.REC.1399.235).
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




