Classic human astrovirus 4, 8, MLB-3, and likely new genotype 5 sublineage in stool samples of children in Nigeria
Abstract
Human astrovirus (HAstV) is a nonenveloped RNA virus and has been implicated in acute gastroenteritis among children and elderly. However, there exists a substantial dearth of information on HAstV strains circulating in Nigeria. Viral-like particles were purified from archived 254 stool samples of children with acute flaccid paralysis between January and December 2020 from five states in Nigeria, using the NetoVIR protocol. Extracted viral RNA and DNA were subjected to a reverse transcription step and subsequent random polymerase chain reaction amplification. Library preparation and Illumina sequencing were performed. Using the virome paired-end reads pipeline, raw reads were processed into genomic contigs. Phylogenetic and pairwise identity analysis of the recovered HAstV genomes was performed. Six near-complete genome sequences of HAstV were identified and classified as HAstV4 (n = 1), HAstV5 (n = 1), HAstV8 (n = 1), and MLB-3 (n = 3). The HAstV5 belonged to a yet unclassified sublineage, which we tentatively named HAstV-5d. Phylogenetic analysis of open reading frames 1a, 1b, and 2 suggested recombination events inside the MAstV1 species. Furthermore, phylogenetic analysis implied a geographic linkage between the HAstV5 strain from this study with two strains from Cameroon across all the genomic regions. We report for the first time the circulation of HAstV genotypes 4, 8, and MLB-3 in Nigeria and present data suggestive for the existence of a new sublineage of HAstV5. To further understand the burden, diversity, and evolution of HAstV, increased research interest as well as robust HAstV surveillance in Nigeria is essential.
1 INTRODUCTION
Human astrovirus (HAstV) is an important cause of diarrheal infection in children and elderly.1, 2 Infection by HAstV is usually subclinical but can present with symptoms consisting majorly of abdominal pains, mild watery diarrhea, and less commonly anorexia, fever, vomiting, headaches, and severe dehydration often leading to hospitalization.1 Astrovirus (Astv) infections associated with nonenteric manifestations including neurological symptoms and encephalitis in immunocompromised patients and in animals have also been documented.3-6 Although fecal–oral transmission among humans believed to be a major mode of transmission, the significant genetic diversity, combined with the potential for recombination events during coinfections with multiple strains, positions Astvs as strong contenders for emerging zoonotic infections.3, 7
The HAstV is about 28–30 nm in diameter with a nonenveloped icosahedral capsid containing a single stranded RNA genome of positive polarity. The genome size ranges between 6.4 and 7.7 Kbp (depending on the species), and is organized into three major overlapping open reading frames (ORFs). Located towards the 5′-end of the genome are ORF1a and ORF1b encoding the nonstructural serine protease and the RNA-dependent RNA polymerase, respectively, whereas on the 3′-end ORF2 encodes the capsid protein. Of note, a fourth ORF designated as ORFX and overlapping with the 5′-end of the capsid protein has been identified recently.8 It encodes the XP protein, which experimental studies have suggested to be essential in virus assembly and/or release.8
HAstVs belong to the family Astroviridae. This viral family contains two genera: Mamastrovirus (MAstV) and Avastrovirus (AVstV) infecting mammals and birds, respectively. Species demarcation criteria in both genera are based on genetic distance between the entire ORF2.9 On average, the amino acid pairwise similarity between the two genera is 17%, and within the MAstV genus a mean similarity of 28% is observed.10 Each Astv genus is divided into genogroups I and II. In the MAstV genus, the mean amino acid genetic similarity between the two genogroups is 33%, and within each genogroup, amino acid homology between genotypes ranges between 21.7% and 66.2%.9 To date, 19 species (Mamastrovirus-1 to Mamastrovirus-19) have been described with only four infecting humans.3 These four include Mamastrovirus-1 consisting of the classical HAstV genotype 1–8 (HAstV1–8), Mamastrovirus-6 containing HAstV clade Melbourne (HAstV-MLB1–3).11 The others include the so-called HAstV-Virginia clades/Human-mink-ovine-like Astv (HAstV-VA/HMO) with VA2/HMO-B and VA4 belonging to Mamastrovirus-8 whereas VA1/HMO-C and VA3/HMO-A are genetically more closely related to Mamastrovirus-9.3, 12 Another species tentatively designated the 20th species and the fifth group infecting human has been proposed.3, 13 The divergence in amino acid sequences across the entire genome between the MLB, VA/HMO, and the classical HAstVs is very high suggesting a possible substantial diversity in their biology and antigenic properties.3 Each genotype (HAstV1–8) of the classical HAstV can further be classified into distinct lineages on the basis of 93%–97% nucleotide sequence similarity in the complete or partial ORF2 region.3, 10 Precisely, parts of the 5′- and 3′-end (designated region C and D, respectively) of the capsid protein have been used in HAstV lineage subclassification.14
Among the classical HAstV genotype, HAstV1 remains the most predominant genotype worldwide but with varying rates of detection across different regions, followed by HAstV2–5 and -8, whereas HAStV6 and -7 are seldomly detected.15 In Africa, the detection of HAstV5 and -8 is more frequent than in other regions.16 In Nigeria, HAstV1 and -5 have been reported by a few studies in children with gastroenteritis.12, 17, 18 The nonclassical HAstVs including the HAstV-MLB-1 and HMO species A and B have also been reported in Nigeria.12 Recently, a yet to be classified HAstV strain closely related to canine Astv was isolated from a Nigerian child with diarrhea.1, 7, 18 Although the role of HAstVs in acute gastroenteritis in children have been established, there exist a substantial dearth of information on the molecular characterization of HAstVs circulating in Nigeria. Also, recombination in Astvs have been reported13, 19-21 in many parts of the world including sub-Saharan Africa, but such occurrence is yet to be investigated in Nigeria. Therefore, leveraging the ongoing national acute flaccid paralysis (AFP) surveillance we explored molecular epidemiology of HAstVs in archived stool samples of Nigerian children (under the age of 15 years) with AFP using viral-like-particle-based metagenomics approach.
2 METHODS
2.1 Sample collection and processing
This was a retrospective descriptive investigation conducted with convenience samples from AFP surveillance program. Fecal samples analyzed in this study were collected in 2020 from children below the age of 15 years diagnosed with AFP in Nigeria. A total of 6330 fecal samples from AFP cases were received by the World Health Organization (WHO) National Polio Laboratory between January and December 2020. All stool samples were collected in accordance with the national ethical guidelines as part of the National AFP surveillance program in Nigeria and sent to the WHO National Polio Laboratory, Nigeria, to ascertain whether poliovirus was the etiologic agent of the diagnosed AFP using the established WHO algorithm.22 Eight of the samples were positive for circulating vaccine-derived poliovirus and none for wild poliovirus.20
Specifically, 254 archived (at −20°C) poliovirus culture-negative fecal samples were randomly selected according to location and month of sample collection from five states in Nigeria representing five geopolitical zones in the country (Lagos; South-west, Anambra; South-east, Edo; South-south, FCT Abuja; North-central and Kaduna; North-west) (Figure 1 and Supporting Information S1: Table 1). After sorting, the samples were anonymized before processing. Thereafter, about 0.5 g of each stool specimen was diluted in 4.5 mL of phosphate buffered saline and 0.5 g of glass beads. The mixture was vortexed for 20 min and thereafter centrifuged at 1469g for 20 min. Subsequently, 2 mL of the supernatant was aliquoted into two cryovials of 1 mL volume each and stored at −20°C. To make a pool, 200 μL of each fecal suspension was added. Pools contained between 1 and 7 fecal suspensions (Supporting Information S1: Table 1).
2.2 Sequencing and reads processing
The pools were processed according to the NetoVIR protocol.21 Briefly, fecal suspensions were centrifuged at 17 000g for 3 min, followed by filtration using a 0.8 μm PES centrifugal filter (Sartorius). To digest free-floating nucleic acids, the filtrate was treated with a cocktail of Benzonase (Millipore) (Novagen) and Micrococcal Nuclease (New England Biolabs) at 37°C for 2 h. Thereafter, nucleic acids were extracted using the QIAamp Viral RNA Mini Kit (Qiagen) according to the manufacturer's instructions. First- and second-strand synthesis and random PCR amplification for 17 cycles were performed using a slightly modified Whole Transcriptome Amplification (WTA2) Kit procedure (Sigma-Aldrich). WTA2 products were purified with MSB Spin PCRapace spin columns (Stratec Biomedical). The sequencing libraries were prepared for Illumina sequencing using the Nextera XT Library Preparation Kit (Illumina). Samples were paired-end sequenced (2 × 150 bp) on an Illumina Novaseq. 6000 platform.
Raw reads were processed with the virome paired-end reads pipeline (https://github.com/Matthijnssenslab/ViPER). Briefly, the reads were trimmed for quality and adapters removal using Trimmomatic23 and reads mapping to the human genome were removed using Bowtie2.24 Subsequently, the trimmed and filtered reads were de novo assembled into contigs using metaSPAdes.25 Contigs were then annotated using DIAMOND with the sensitive option.26 The trimmed and filtered reads were also de novo assembled using MEGAHIT on the KBase platform using default parameters.27 Contigs were then annotated using a BLASTn search against the GenBank database. Identified AstV contigs recovered from both assembly pipelines were merged and de-duplicated. Subsequently, to obtain depth of coverage, trimmed reads were mapped against the AstV contigs using Bowtie2.24
2.3 Phylogenetic analysis
The AstV contigs recovered in this study were used as query in a BLASTn search of the GenBank database. Top hits were downloaded and used to make a local database to which sequences recovered in this study were added. In all, 67 complete HAstV reference genomes (including representative sequences of classical HAstV1–8 and MLB1–3) were retrieved, and altogether 73 genome sequences (including six near-complete sequences from this study) were included in subsequent analyses. This database of near-complete HAstV genomes was subjected to a Multiple Sequence Alignment using ClustalW in MEGA 11 software.28 Using ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/) individual ORFs were mapped. Different genomic regions of interest were selected from the alignment and used to make corresponding maximum-likelihood phylogenetic trees (GTR model and 1000 bootstrap replicates) in MEGA 11 software.28 Precisely, the genotypes of all HAstV strains and sub-classification into distinct lineages for classical HAstV (MAstV1) obtained from this study were inferred from phylogenetic trees made from the complete ORF2 region. To further clarify lineage sub-classification for one of the classic HAstV phylogenetic trees of complete and partial {5′ (region C) and 3′ (region D) ends} ORF2 regions were drawn in parallel. This was also subjected to pairwise identity analysis using SDT version 1.2.29 Varying phylogenetic topologies across the three major ORF regions in HAstV were accessed for ORF1a, ORF1b and ORF2 regions extracted from the near-complete genome alignment. To explore the inconsistency observed in the tree topology across the three major ORFs in MAstV-1, selected sequences were used to construct individual phylogenetic trees as earlier described. To establish the recombination event in one of the MAstV-1 isolated from this study, bootscan and similarity plot was implemented on SimPlot 3.5.1 software (https://sray.med.som.jhmi.edu/SCRoftware/SimPlot/) Bio-edit sequence alignment editor was used to view the predicted amino acid sequence in the ORF-X region of the study HAstV strains.
To understand the evolutionary relationship between the MLB strains from this study and similar genotypes described in other sub-Saharan African countries,30 we assembled a second database containing the MLB sequences described in this study alongside partial ORF1b sequences of 25 other MLB-3 strains circulating in Gambia and Kenya that had been deposited in GenBank. The sequences in this database were analyzed as described above.
2.4 GenBank submission
The sequences and mapped reads described in this study have been submitted to GenBank and the SRA and have been assigned accession numbers OR448830–OR448835 and PRJNA1004047, respectively.
3 RESULTS
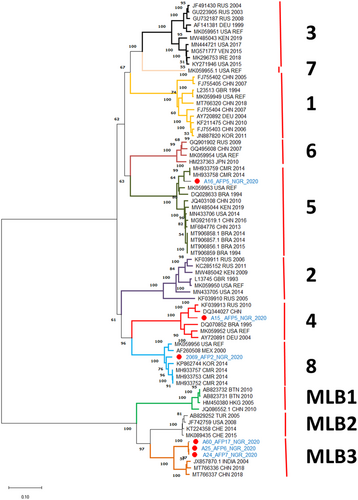
Six Astv near-complete genomes (2068_AFP2_NGR_2020, A15_AFP5_NGR_2020, A16_AFP5_NGR_2020, A25_AFP6_NGR_2020, A24_AFP7_NGR_2020, and A60_AFP17_NGR_2020) were recovered. The GC content, mean coverage and mapped reads range from 40% to 44%, 78× to 9375×, and 3595 to 384 815, respectively (Table 1). BLASTn search indicated close relationships with HAstV genotypes HAstV4 (n = 1), HAstV5 (n = 1), HAstV8 (n = 1), and MLB-3 (n = 3) (Table 2). These classifications were confirmed in a phylogenetic analysis based on the complete ORF2 region of the six HAstV genomes detected in this study (Figure 2).
| SAMPLE IDa | Length | GC (%) | Mean coverage | Mapped reads | Total reads |
|---|---|---|---|---|---|
| 2069_AFP2_NGR_2020 | 6843 | 44 | 78.8 | 3595 | 11 361 362 |
| A15_AFP5_NGR_2020 | 6789 | 43 | 197.8 | 8964 | 5 775 766 |
| A16_AFP5_NGR_2020 | 6735 | 44 | 102.3 | 4592 | 5 775 766 |
| A25_AFP6_NGR_2020 | 6152 | 40 | 747.1 | 30 600 | 5 742 760 |
| A24_AFP7_NGR_2020 | 6145 | 40 | 556.3 | 22 739 | 3 754 582 |
| A60_AFP17_NGR_2020 | 6160 | 40 | 9375.9 | 384 815 | 10 867 948 |
- Abbreviations: AFP, acute flaccid paralysis; HAstV, human astrovirus.
- a Sample ID (2069_AFP2_NGR_2020) means contig 2069 from pool 2 collected in Nigeria in year 2020).
| Hit 1 | Hit 2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| S/N | ID | Accession # | HAstV type | Accession # | Pairwise identity (%) | Country (year) | Accession # | Pairwise identity (%) | Country (year) |
| 1 | 2069_AFP2_NGR_2020 | OR448830 | HAstV-8 | KP862744 | 96.77 | South Korea (2014) | MH933753 | 96.20 | Cameroon (2014) |
| 2 | A15_AFP5_NGR_2020 | OR448831 | HAstV-4 | KF039912 | 96.02 | Russia (2005) | MT906853 | 95.62 | Brazil (2014) |
| 3 | A16_AFP5_NGR_2020 | OR448832 | HAstV-5 | MH933759 | 95.68 | Cameroon (2014) | MH933758 | 95.68 | Cameroon (2014) |
| 4 | A25_AFP6_NGR_2020 | OR448833 | MLB-3 | JX857870 | 90.93 | India (2004) | MT766337 | 90.46 | China (2018) |
| 5 | A24_AFP7_NGR_2020 | OR448834 | MLB-3 | JX857870 | 90.92 | India (2004) | MT766337 | 90.45 | China (2018) |
| 6 | A60_AFP17_NGR_2020 | OR448835 | MLB-3 | JX857870 | 90.83 | India (2004) | MT766337 | 90.32 | China (2018) |
- Abbreviation: HAstV, human astrovirus.
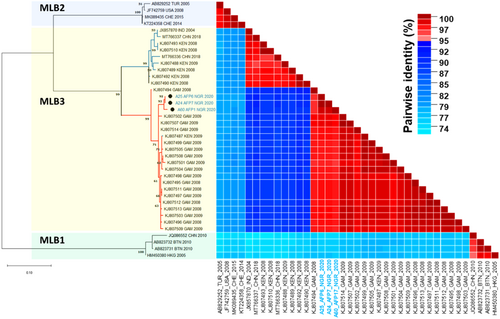
The HAstVs MLB1–3 (belonging to species Mamastrovirus 6) are genetically distinct from the classical HAstV1-8 (belonging to species Mamastrovirus 1). Phylogenetic analysis of the partial ORF1b (comprising 408 bps position 3109–3517 JX857870) of the MLB-3 isolated in this study and similar genotypes reported28 from Kenya and Gambia between 2008 and 2009 showed that the strains from Kenya and Gambia formed two distinct clades with our Nigerian isolates clustering more closely with the Gambian strains (Figure 3). Pairwise identity analysis showed nucleotide divergence between these two clusters ranging between 9% and 12%, whereas divergence within clusters ranged between 0.7% and 3% (Figure 3). Amino acid conservation analysis showed ORF1b substitution of asparagine at position 356 by aspartic acid (N356D) in all MLB-3 sequences of the Gambia/Nigeria cluster (Supporting Information S2: Figure S1).
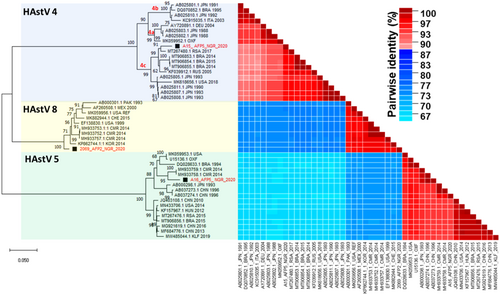
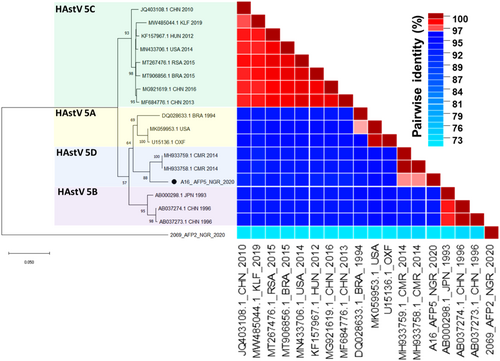
Molecular analysis of the entire ORF2 as well as small portions of the 5′ (region C) and 3′ (region D) ends of ORF2 have shown distinct sequence variations within MAstV-1 enabling further genetic lineage distinction. Classification of some members of MAstV-1 from this study into their sublineages using the complete ORF2 region as described previously31 showed that our HAstV4 isolate belongs to sub-lineage 4c with >94% pairwise identity with other 4c strains (Figure 4). The HAstV5 genome recovered from the study clustered most closely with two strains detected in 2014 in Cameroon (MH933758 and MH933759), which currently fall outside the established HAstV5 sublineages a–c (Figure 5). Further exploration of this subcluster indicated a possible new HAStV-5 sublineage d with ≥96% similarity in the nucleotide sequences of the complete ORF2 within this cluster (Figure 5 and Supporting Information S1: Tables S2 and S3). Same clustering pattern was noted within this lineage even when partial ORF2 regions (the C and D regions position 4526–4974 and 6477–6715 of L13745, respectively) were used for the sublineage classification (Supporting Information S1: Figure S2).
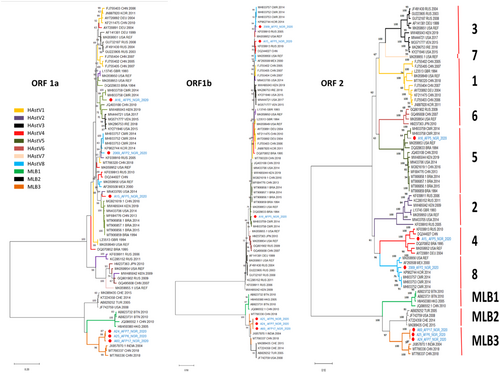
Recombination is one of the drivers of evolution in RNA viruses and can affect phylogenetic grouping. Typically, in HAstV recombinants display varying topologies across the different ORFs.19, 31 Phylogenetic trees (Figure 6) generated in parallel for ORF1a, ORF1b, and ORF2 to assess for varying phylogeny topologies across the three major ORF regions showed consistency in the topology of the MLB-3 sequences but inconsistency in tree topology of the three ORFs for MAstV1 (classical HAstV). Separate phylogenetic trees constructed with selected MAstV-1 sequences showed that in ORF1a the HAstV-4 detected in this study (A15_AFP5_NGR_2020) clustered with a genotype 2 strain isolated in 2014 from the USA (MN433705) with a bootstrap support of 93% (figure not shown). Similarity plot and bootscan analysis with A15_AFP5_NGR_2020 as query alongside the strain from the USA (MN433705) and two other reference strains (OP4322301; HAstV-4 and AF141381; HAstV-3) showed that recombination events contributed to the emergence of A15_AFP5_NGR_2020 (Supporting Information S2: Figure S3). Also, although the HAstV-5 detected in this study consistently clustered with the two Cameroonian isolates (MH933758 and MH933759), these three west African strains failed to cluster with other HAstV-5 strains in the ORF1a and ORF1b genomic regions (Figure 6).
The newly identified ORFX encodes the XP protein, which is thought to be essential for virus assembly and/or release.8 It is reported to typically range between 70 and 112 amino acids. We identified this ORF in our genomes, and in the MLB-3 isolates it is located about 41 nucleotide downstream of the ORF2 start codon and is made up of 204 nucleotides translating into a protein product of 67 amino acids (Supporting Information S2: Figure S3). In MAstV1 (HAstV4, 5, and 8), this ORFX consisted of 399 nucleotides (starting between 44 and 47 nucleotides downstream of the ORF2 start codon) translated in +1 reading frame resulting in a protein of 112 amino acids (Supporting Information S2: Figure S4).
4 DISCUSSION
There is scarce data from Nigeria regarding the evolution and molecular characterization of Astv viruses. In this study, we detected six near-complete genomes of HAstV belonging to the classical genotypes HAstV4, HAstV5, and HAstV8, as well as three closely related MLB-3 strains from stool samples collected from children with AFP in Nigeria. Notably, except for HAstV5, no prior sequence data existed in GenBank for any of the genotypes detected (MLB-3, HAstV4, and HAstV8) in Nigeria. In addition, this study reported the first MLB-3 near-complete genome sequence from sub-Saharan Africa and deposited into the GenBank. It is our opinion that these data may not necessarily infer new introductions of these genotypes into Nigeria, but rather reflects a suboptimal surveillance for HAstVs in Nigeria. Of note, with the recent roll out of Rotavirus vaccination into the routine childhood immunization schedule in Nigeria,32 other viruses like Astv and norovirus may have covertly continued to play a more clinically significant role in the etiology of pediatric viral gastroenteritis in the country. This calls for an increased interest in these viruses to better understand their biology, epidemiology, and clinical significance.
Since its first report in 2013 in India,33 the MLB-3 genotypes have now been described in other regions including China,34 Japan (unpublished), Kenya, and Gambia.30 Previously, MLB-1 have been described in Nigeria12 although sequences are not yet available in GenBank. As of now MLB-2 has yet to be reported in Nigeria. Of interest was the observation that both of the identified MLB-3 strains were found in samples from Anambra, potentially suggesting local circulation of this strain in the region and/or a local outbreak. Based on the complete ORF2, the MLB-three genomes described in this study formed a separate lineage with 100% bootstrap support from the only other available complete MLB-3 genomes from India (JX857870) and China (MT766337 and MT766336) (Figure 2). Although ICTV recommends criteria for Astv classification to be based on the amino acid pairwise distances of the complete ORF2, the so-called regions A, B, C, and D in the ORF1a, ORF1b, and 5′- and 3′-ends of ORF2 (according to Martella et al.14), respectively, have also been used by other studies for virus classification.14, 35, 36 Indeed the partial ORF1b region (region B) of HAstV-MLB-3 was used to classify the other MLB-3 strains isolated from sub-Saharan Africa by Meyer et al.30 Phylogenetic analysis (partial ORF1b region, 408 bp; position 3109–3517 using JX857870 as the reference) showed that MLB-3 detected in this study (although forming a separate lineage) shared a common ancestor with all Gambian isolates and one other from Kenya isolated in 2008 and 2009, whereas the rest of Kenyan isolate clustered with the Chinese and Indian strains with 99% bootstrap support (Figure 3). The MLB-3 clustering pattern in Figure 3 suggests a geographic clustering pattern. Although more MLB-3 sequence data of preferably the entire ORF2 from across the region and other sub-Saharan African nations will be essential to ascertain a clear geographical MLB-3 clustering pattern. This will be useful for molecular epidemiology.
Classical HAstVs contigs detected in this study were classified as genotypes HAstV4, HAstV5, and HAstV8 based on the complete ORF2 nucleotide sequence (Figure 2). The HAstV4 sequence described in this report is currently the only one from both West and Central Africa and the second from sub-Saharan Africa available in GenBank. The HAstV4 from this study share a common ancestor with HAstV4 circulating in China and Russia, suggesting wider distribution of this genotype. Again, to the best of our knowledge we describe here the first complete coding sequence data of HAstV5 from Nigeria. The HAstV5 detected in this study belongs to the same lineage as those circulating in Cameroon, a country that shares borders with both the Norths and Southern part of Nigeria. Although HAstV5 have earlier been described in Nigeria by Arowolo et al. in 2020,17 we could not include their strains in our analysis because the authors only amplified partial sequences at the 5′-end of ORF2 (256 bp out of the 448 bp of the C region). The HAstV8 genome clustered with other HAstV8 strains from other parts of the world including neighboring Cameroon (Figure 2) with 100% bootstrap support. It is documented16 that HAstV-5 and -8 are more commonly isolated in Africa than in other regions of the world.
Several studies have tried to systematically classify the genotypes HAstV1–8 into distinct sublineages according to the nucleotide divergence of either the complete genome14 or portions of the capsid proteins.37, 38 Martella et al.14 identified six lineages within HAstV1 (1a–1f), four within HAstV2 (2a–2d), two within HAstV3 (3a and 3b), three within HAstV4 (4a–4c), three within HAstV5 (5a–5c), two within HAstV6 (6a and 6b), and one for HAstV 7 and 8. Recently, a third sublineage of HAstV3 designated 3c has been proposed.39 For genotype HAstV4, inter-sublineage nucleotide similarities range from 89.1% to 93.5% in the entire ORF2 region, whereas intralineage nucleotide similarity of at least 94.3% were observed14 Consequently, based on a 94% nucleotide similarity cut-off our HAstV4 genome was subclassified as genotype 4c. Although, specific temporal or geographical pattern have not been demonstrated in genotype 4, HAstV-4c have been shown to have a wider geographic spread than other subtypes.31
Furthermore, subclassification based on the entire ORF2 region showed that HAstV5 from this study segregated very closely and independently (with very high bootstrap support) with some yet to be subclassified HAstV5 (MH933758 and MH933759) strains isolated in 2014 from neighboring Cameroon (Figure 4).40 These three strains showed ≥96% nucleotide similarity in the complete ORF2 (Figure 4). Notably, partial sequences of other regions of ORF2 such as region C and D have been used for subclassification of MAstV1.14, 31, 41, 42 We observed similar clustering pattern among the entire ORF2 region and the partial regions (C: position 4526–4974; D: position 6477–6715 of L13745), although with a lower statistical support in region D (bootstrap value < 50%) (Supporting Information S2: Figure S2). Although inconsistencies have been observed in lineage clustering pattern between trees drawn from complete ORF2 CDS and region D with the latter having lower statistical support for subclassification,14 the HAstV-5 detected in this study and two others from neighboring Cameroon showed a consistent topology across all the regions, suggesting that this lineage may be novel. Based on these tree topologies, strong bootstrap support and pairwise identity data, we tentatively refer to this as sublineage HAstV5d (Figure 5). Nevertheless, other independent report preferably with more HastV5 sequences displaying similar clustering pattern will be necessary to confirm our findings and deliver needed supporting data for designation of this potential new lineage HAstV5d (Figure 5). Particularly, Martella et al.14 suggested that novel lineages should only be assigned if a complete ORF2 sequence is available for the potential new strain and at least two other independent reports showing epidemiological relevance of the strain. We therefore await other reports to further confirm and possibly shed more light on the epidemiological relevance of the new strain.
Recombination is a common event among MAstV-1 strains with recombinants often displaying phylogenetic discrepancies across the different ORFs.20, 32 The topology of our trees and Simplot analysis with bootstrap support suggests that recombination events contributed to the emergence of A15_AFP5_NGR_2020 and/or the lineage to which it belongs (see Figure 6 and Supporting Information S2: Figure S3). Further, the tree topologies suggest a geographic linkage especially in genotype 5 where the HAstV5 from this study continued to cluster with the two strains from Cameroon (recovered from neighboring countries sharing common borders) across all the three ORF regions (Figure 6). More reports of HAstV5 from the region will be necessary to fully understand the clustering pattern observed in our study. The presence of ORFX overlapping the 5′-end of ORF2 in both Mamastrovirus-1 (HAstV1–8) and Mamastrovirus-6 (MLB) reported in this study confirmed as predicted by Lulla et al.8 the existence of this ORF even in divergent MLB strains. In their study they suggested that the ORFX encodes the XP protein, which typically ranges from 70 to 112 amino acid and is predicted to be involved in virus assembly and release.8 We noted that the protein product (XP protein) of the ORFX in MLB-3 and classical HAstV8, 4, and 5 strains isolated in this study had 67 and 112 amino acids, respectively (Supporting Information S2: Figure S4). Amino acid variations were common among the classical HAstV (Mamastrovirus-1) from this study. However, due to limited knowledge of the function and three-dimensional structure of this protein, it is not possible to speculate about the effect of amino acid changes on the function of the protein and the fitness of the virus.
In summary, we report for the first time the circulation of genotypes HAstV4, HAstV8 and the divergent MLB-3 with a possible geographic pattern of distribution, and present data suggesting presence of a new sub-class of HAstV-5 in Nigeria.
AUTHOR CONTRIBUTION
Sample selection and processing: George E. Uwem, Agbaje T. Sheriff, Olayinka A. Oluseyi, Oni I. Elijah, Akinleye E. Toluwanimi, Popoola O. Bolutife, Olayinka O. Titilola, and George A. Oluwadamilola. Study design: Ifeorah M. Ijeomah, Faleye O. C. Temitope, George E. Uwem, Adeniji A. Johnson, Matthijnssens Jelle, and Adewumi M. Olubusuyi. Laboratory analysis: De Coninck Lander and Matthijnssens Jelle. Data analysis: De Coninck Lander, Ifeorah M. Ijeomah and Faleye O. C. Temitope. Writing—initial draft of the manuscript: Ifeorah M. Ijeomah and Faleye O. C. Temitope. Writing—review and editing: De Coninck Lander, Agbaje T. Sheriff, George E. Uwem, Onoja A. Bernard, Olayinka A. Oluseyi, Ajileye G. Toluwani, Oragwa O. Arthur, Osasona G. Damilola, Ahmed I. Muhammad, Komolafe I. Omotosho, Adeniji A. Johnson, Matthijnssens Jelle and Adewumi M. Olubusuyi; Supervision: Adeniji A. Johnson, Matthijnssens Jelle and Adewumi M. Olubusuyi. All the authors have read and approved the final version of the manuscript before submission.
ACKNOWLEDGMENTS
We thank the WHO National Polio Laboratory in Ibadan, Nigeria for providing archived stool samples analyzed in this study. There was no funding for the study.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ETHICS STATEMENT
Stool samples analyzed in this study were collected in accordance with the national ethical guidelines as part of the National AFP surveillance program in Nigeria. The samples were sent to the WHO National Polio Laboratory in Ibadan, Nigeria, to ascertain whether poliovirus is the etiologic agent of the diagnosed AFP using the WHO algorithm. The samples were anonymized before use in the study; thus, the article does not contain any information that can be used to associate samples analyzed to any individual.
Open Research
DATA AVAILABILITY STATEMENT
Sequence data presented in this manuscript have been submitted to GenBank (accession numbers OR448830–OR448835) and the SRA (accession number PRJNA1004047).




