Nationwide epidemiologic and genetic surveillance of hepatitis E in Japan, 2014–2021
Abstract
Hepatitis E virus (HEV) is an emerging causative agent of acute hepatitis. To clarify the epidemiology of HEV and characterize the genetic diversity of the virus in Japan, nationwide enhanced surveillance and molecular characterization studies of HEV in Japan were undertaken from 2014 to 2021. In total, 2770 hepatitis E cases were reported, of which 88% were domestic cases, while only 4.1% represented cases following infection abroad. In addition, 57% of domestic infections occurred in males aged in their 40s–70s. For domestic cases, infection via pork meat consumption continued to be the most reported route. Analysis of the 324 sequences detected between 2016 and 2021 showed that the majority of domestic HEV strains belong to Genotype 3a (G3a) and G3b. In contrast, six of eight cases of G1 HEV reflected infection abroad. Our results suggest that HEV is circulating widely in Japan, with genotypes G3a and G3b being most prevalent. Continued surveillance is necessary to monitor future trends and changes in the epidemiology of HEV in Japan.
Abbreviations
-
- HEV
-
- hepatitis E virus
-
- MHLW
-
- Ministry of Health, Labour and Welfare, Japan
-
- NESID
-
- National Epidemiological Surveillance of Infectious Diseases
-
- NIID
-
- National Institute of Infectious Diseases
1 INTRODUCTION
Hepatitis E is an acute hepatitis caused by infection with hepatitis E virus (HEV), a member of the Hepeviridae family; this virus is the most common cause of acute viral hepatitis, with an estimated 20 million or more cases occurring worldwide each year.1 Hepatitis E has a long incubation period of 15–60 days and is associated with fever, general malaise, nausea, vomiting, anorexia, abdominal pain, and jaundice; however, subclinical infection is also common. Although HEV infection is not conventionally thought to be chronic, HEV infection in immunosuppressed patients, such as organ transplant recipients, can cause chronic infection. HEV that infect humans belong to the taxon Paslahepevirus balayani (formerly Orthohepevirus A), a subfamily of Orthohepeviridae. P. balayani is further classified into eight genotypes (G1–G8), and each genotype is separated further into several subtypes.2
HEV G1 to G4 are the major genotypes known to infect humans. HEV G1 and G2 are human pathogens that are prevalent mainly in developing countries; these genotypes are transmitted via the fecal–oral route, causing waterborne outbreaks. In contrast, HEV G3 and G4 are zoonotic pathogens with a broad host range that includes swine, wild boar, and other animal species; these genotypes sporadically result in cases of hepatitis E in developed countries, including Japan.3-6 Chronic hepatitis E is most commonly reported in immunosuppressed patients infected with HEV G3, the prevalent genotype infecting humans in developed countries. In addition, it has also been reported that HEV G4 and G7 can cause chronic infections in transplant recipients.7, 8
The number of reported cases of acute hepatitis E in Japan has continued to increase since coverage for the anti-HEV-IgA detection assay kit was initiated by the National Health Insurance System in October 2011. Hepatitis E is classified as a category IV infectious disease under the Infectious Diseases Control Law in Japan, and the government must be notified of all diagnosed cases. In addition, the Ministry of Health, Labour and Welfare, Japan (MHLW), requests local governments to collect patient specimens for molecular epidemiological analysis and to cooperate in active surveillance as of 2016 (August 16, 2016: Administrative notice #0816-3 and Raw Food Supervision #0816-2). Since then, molecular epidemiological analysis has been conducted by the sequencing of viral genomes recovered from patient stool or serum specimens; this analysis is conducted in collaboration with prefectural and municipal public health institutes (PHIs) and the National Institute of Infectious Diseases (NIID) in an effort to investigate the commonality of infection sources. In the present surveillance report, we present an overview of the epidemiological characteristics of the reported hepatitis E cases diagnosed between 2014 and 2021 in Japan; our analysis includes distribution by age, sex, and geography, as well as molecular epidemiologic profiles.
2 MATERIALS AND METHODS
2.1 Data collection
The National Epidemiological Surveillance of Infectious Diseases (NESID) system has been operating under the Infectious Diseases Control Law in Japan since 1999 (https://www.niid.go.jp/niid/images/epi/nesid/nesid_en.pdf). Physicians are required to notify public health centers (PHCs) of all notifiable diseases (the reporting criteria and the notification form for each disease are publicly available), and these centers then coordinate appropriate responses with prefectural and municipal PHIs. Laboratory confirmation methods accepted for public health surveillance for Hepatitis E include the detection of HEV RNA by polymerase chain reaction (PCR) in patient blood and/or stool samples, or the detection of either anti-HEV immunoglobulin M (IgM) or anti-HEV IgA in patient serum. The notification form includes demographic, clinical, laboratory, and exposure information. The data collected by the PHCs and PHIs are then reported using the electronic NESID system. Hepatitis E cases diagnosed between 2014 and 2021 were extracted from the NESID database on October 26, 2021. This surveillance study was conducted based on the NESID system using data collected for public health purposes (https://www.mhlw.go.jp/content/10900000/000488981.pdf) and did not require informed consent or ethical approval.
2.2 Sequencing of HEV RNA
Available serum and/or stool specimens from patients with hepatitis E were submitted to the prefectural and municipal PHIs or NIID with no screening. Generally, viral RNA was extracted from 140 µL of serum sample or stool suspension, as described previously.9 A 506-bp or 457-bp partial genome fragment from the HEV open reading frame 2 (ORF2) region was amplified by nested reverse transcription (RT) -PCR with the primer pair HE044 (5′-CAAGGHTGGCGYTCKGTTGAGAC-3′) and HE040 (5′-CCCTTRTCCTGCTGAGCRTTCTC-3′) in the first round and HE110-2 (mixture of three sequences, 5′-GYTCKGTTGAGACCTCYGGGGT-3′, 5′-GYTCKGTTGAGACCACGGGYGT-3′, and 5′-GYTCKGTTGAGACCTCTGGTGT-3′) and HE041 (5′-TTMACWGTCRGCTCGCCATTGGC-3′) in the second round.10 The amplified PCR products were purified using the Wizard SV Gel and PCR Clean-Up System (Promega), followed by Sanger sequencing.
2.3 Phylogenetic analysis
The 412 nucleotides of the amplified partial HEV ORF2 sequence, excluding the primer sequence, were aligned with reference HEV sequences of various genotypes and subtypes.2 Phylogenetic analysis was conducted using the MEGA program (version 10.2.6) and the neighbor-joining method. The reliability of the tree at each branch node was assessed by the bootstrap method with 1000 replicates. The nucleotide sequence data used in this analysis are available from the authors upon request.
3 RESULTS
3.1 Demographic characteristics of hepatitis E in Japan from 2014 to 2021
A total of 2770 hepatitis E cases was diagnosed between January 2014 and September 2021 and reported via the NESID system11 (Figure 1A). The majority of patients (88%) was reported as infections presumed to be acquired endemically (i.e., domestic cases), and the presumed number of infections abroad was only 4.1%. The reported place of infection (domestic vs. abroad) was unknown/undetermined for the remaining cases (7.9%).
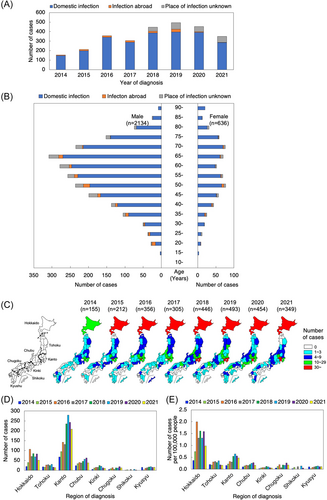
The majority of reported HEV infections were male cases, representing 2134 cases (domestic infection: 1865 cases; infection abroad: 90 cases; place of infection unknown: 179 cases); in contrast, there were 636 female cases (domestic infection 567: cases; infection abroad: 23 cases; place of infection unknown: 46 cases) (Figure 1B). Regarding the age distribution of HEV infection, most patients were of middle age or older for both sexes. Specifically, males in their 40s–70s were the most reported cases, accounting for 57% of domestic infections.
Geographically, many hepatitis E cases were reported from Hokkaido and the Kanto region, and relatively few from the Shikoku region (Figure 1C,D, distribution of the 2770 cases by prefecture). The Kanto region has the highest number of reports, but Hokkaido stands out in terms of cases per population (Figure 1E). The main reported (presumed) place of infection in 113 cases following infection abroad was Asia, with China having the highest number (20%), followed by India (15%), Taiwan (7.1%), and Thailand (7.1%) (Table 1).
| Infected abroad cases | 113 |
|---|---|
| China | 23 |
| India | 17 |
| Taiwan | 8 |
| Thailand | 8 |
| United States of America | 7 |
| South Korea | 6 |
| Pakistan | 5 |
| The Philippines | 4 |
| Malaysia | 2 |
| Vietnam | 2 |
| Bangladesh | 2 |
| Germany | 2 |
| Singapore | 2 |
| Australia | 2 |
| Other | 9 |
| Two or more countries | 7 |
| Place of infection unknown | 7 |
3.2 Suspected routes of infection
Of the 2770 diagnosed cases from 2014 to 2021, information regarding the reported infection route was available for 1035 cases in which domestic infection was presumed/suspected. Among these cases, 428 (41%) reported recent consumption of pork (including liver for some cases); another 99 (10%) wild boar meat, and 88 (9%) deer meat (Table 2; note that some cases reported more than one of these and the figures are not mutually exclusive). Among the 113 cases in which infection abroad was suspected, 30 (27%) were attributed to the consumption of pork or the meat of unidentified animals, and 5 (4%) were attributed to drinking water. For all of these data, it should be noted that the recorded infection routes are presumed based on patient reports, and not confirmed.
| Domestically infected cases | n = 1035 n (%) |
|---|---|
| Pork | 428 (41) |
| Wild boar meat | 99 (10) |
| Deer meat | 88 (9) |
| Infected abroad cases | n = 113 n (%) |
|---|---|
| Consumption of pork or meat of unidentified animals | 30 (27) |
| Drinking of water | 5 (4) |
- Abbreviation: HEV, hepatitis E virus.
3.3 Genotyping
Among the 2485 cases reported from 2016 to 2021, 324 viral sequences of the partial ORF2 region were obtained. Based on these sequences, the genotypes of these 324 cases were classified as G1 (n = 8 cases), G3 (n = 300), and G4 (n = 16) (Table 3). No G2 isolates were identified. Among the eight G1 isolates, six (75%) were considered to be the result of infection abroad. Of these six cases, five and one appeared to have occurred in India and Bangladesh, respectively (Table 4). In contrast, 64% (193 of 300) and 63% (10 of 16) of the G3 and G4 infections, respectively, appeared to be the result of domestic infections.
| Year | Genotype 1 | Genotype 2 | Genotype 3 | Genotype 4 | Total |
|---|---|---|---|---|---|
| 2016 | 0 | 0 | 11 | 1 | 12 |
| 2017 | 3 | 0 | 19 | 0 | 22 |
| 2018 | 3 | 0 | 72 | 5 | 80 |
| 2019 | 2 | 0 | 84 | 4 | 90 |
| 2020 | 0 | 0 | 61 | 0 | 61 |
| 2021 | 0 | 0 | 53 | 6 | 59 |
| Total | 8 | 0 | 300 | 16 | 324 |
- Abbreviation: HEV, hepatitis E virus.
| Genotype 1 | Genotype 3 | Genotype 4 | Total | |
|---|---|---|---|---|
| Domestically infected cases | 1 | 193 | 10 | 204 |
| Not determined | 1 | 103 | 4 | 108 |
| Infected abroad cases | 6 | 4 | 2 | 12 |
| India | 5 | 0 | 1 | 6 |
| Bangladesh | 1 | 0 | 0 | 1 |
| Indonesia | 0 | 1 | 0 | 1 |
| Singapore | 0 | 1 | 0 | 1 |
| Germany | 0 | 1 | 0 | 1 |
| Taiwan | 0 | 1 | 0 | 1 |
| China | 0 | 0 | 1 | 1 |
- Abbreviation: HEV, hepatitis E virus.
3.4 Phylogenetic analysis
All 324 viral sequences of the partial ORF2 region were subjected to phylogenetic tree analysis using the neighbor-joining method. As shown in Figure 2A, 38% of the sequences clustered with the reference sequence of G3 subtype a (G3a), and 45% of the sequences clustered with the reference sequence of G3b, suggesting that the majority of HEV strains in Japan belong to subtypes 3a and 3b. Eleven sequences clustered with the reference sequence of G3k, a new potential subtype, which has been identified in humans and pigs in Japan.12 Nine sequences clustered with the reference sequence of G3e, which has been identified in wild boar in Mie Prefecture in Japan and is thought to have been introduced from Europe into Japan through the importation of pigs in the 1960s.13 Five sequences clustered with the reference sequence of G3f, which is present mainly in Europe and Thailand. However, G3f has also been isolated from Japanese acute hepatitis E patients without a history of travel abroad, leading to the suggestion that this genotype may have entered Japan through imported pork meat.14 Three sequences clustered with the reference sequence of G3c, which is circulating in Europe.15 Three sequences were unclassified (G3uc) in our study, and clustered separately from G3b. None of the sequences obtained in the present study clustered with the reference sequence of G3ra.
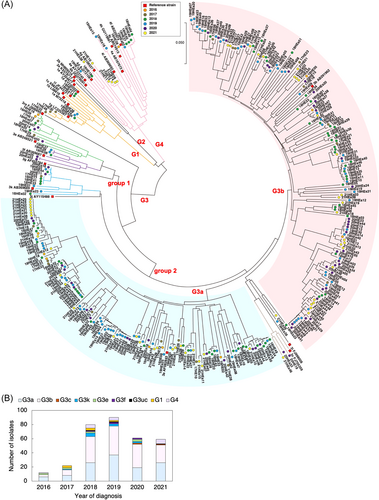
Sixteen sequences clustered with the reference sequence of G4, a genotype that has been isolated from human and pig exclusively in countries of East and Southeast Asia, including Japan.16 Among the eight sequences classified as G1, six sequences clustered with the reference sequence of G1g.
4 DISCUSSION
The present study sought to clarify the occurrence and diversity of HEV in Japan. Before 2011, the annual reported number of hepatitis E cases in Japan was less than 70.17 However, that number has continued to increase since 2012, and more than 300 cases have been reported annually since 2016 (Figure 1A). This increasing trend might reflect an increase in awareness and testing, given that the National Health Insurance System initiated coverage of an anti-HEV-IgA detection assay kit since October 2011. Notably, the overall characteristics of hepatitis E cases in Japan, such as sex, age, geographical distribution, and major genotypes do not appear to have changed. For example, the present study found that older males were at elevated risk of hepatitis E, which is consistent with previous studies conducted in Japan17 and in other countries.18 However, it remains unclear why sporadic acute hepatitis E occurs primarily among older males. Further investigation will be needed to address this observation.
Our analysis of the geographical distribution of HEV infection revealed that Hokkaido and the Kanto region exhibited high numbers of reported cases in Japan (Figure 1C). The Japanese Red Cross Society initiated HEV screening of donated blood in Hokkaido in 2005, because Hokkaido was considered to be a highly endemic area for HEV.19 Thereafter, nationwide HEV-NAT (nucleic acid testing) screening of all donated blood was launched in 2020 to ensure the safety of blood transfusions. Since then, there has been a slight increase of reported hepatitis E cases due to blood screening tests (data not shown), but the proportion is not high and the impact on the total number of cases appears limited (only those who develop symptoms/signs are requested to seek medical attention which leads to notification). Our analysis of the sequences of HEV isolated in the present study revealed that among patients with hepatitis E, G3 is the most prevalent genotype in Japan (Table 3). Considering the high prevalence of HEV G3 in pig,9, 20 a zoonotic food-borne route is presumed to be the primary route of HEV infection in Japan. Wild boar is also known as a reservoir for HEV, although the prevalence of HEV infection in wild boar is lower than that in pig.21 However, HEV RNA prevalence in pigs sold at 6 months of age is not high.9, 22 In addition, pork is generally cooked before ingestion to avoid food poisoning by microbes such as Salmonella, Campylobacter, and other potential pathogens. Nonetheless, the suspected route of 41% of domestic HEV infection cases was suspected to be due to the consumption of pork (Table 2). Given that the consumption of raw or undercooked pork or wild boar products is a risk factor for HEV infection,23 consumers are recommended to cook meat products thoroughly, especially pork and wild boar as HEV has been shown to be inactivated by heating (cooking) to appropriate temperatures.24
Previous investigations of the molecular epidemiology of HEV G3 showed that the virus can be grouped into several subtypes and three monophyletic clades: group 1 (subtypes HEV-3e, f and g); group 2 (HEV-3a, b, c, h, i, j, k, l and m); and HEV-3ra, for which the main host is rabbit.2, 18 A phylogenetic analysis based on partial ORF2 sequences revealed that, among 300 cases classified as G3 in the present study, 94.3% were categorized as members of HEV G3 group 2. Only 14 isolates were categorized as members of HEV G3 group 1 which, compared with HEV G3 group 2, is known to be associated with a more severe course of disease that often results in hospitalization and death.25 Notably, none of the obtained sequences were categorized as HEV-3ra. Among the isolates that were categorized as members of HEV G3 group 2, the majority of the HEV strains isolated in Japan belonged to subtypes 3a and 3b, including 122 (43.3%) that clustered with G3a and 146 (51.8%) that clustered with G3b. In a previous study, G3b was reported to be the main subtype detected in Japan13; however, the present study revealed that G3a is also a common subtype of HEV in Japan.
HEV G4 has also been isolated from human and pig in Japan, China, and Taiwan.16, 26 From 2016 to 2021, a total of 16 HEV isolates were assigned to subtypes 4f, 4i, 4a, 4d, and 4g in the present study. Notably, a sequence obtained from a patient who had traveled to China before the onset of the disease was categorized as 4d.
While a small number of sequences that were categorized as HEV G1 were detected from 2017 to 2019, no G1 sequences were identified among isolates obtained in 2020 and 2021. Presumably, travel restrictions due to the coronavirus disease 2019 (COVID-19) pandemic reduced the number of HEV G1 infections in Japan, given that most G1 cases identified in the present study reflected infections acquired abroad (Table 4). Among the eight sequences categorized as G1, six sequences clustered with the reference sequence of G1g, and five of these six sequences were obtained from patients who appeared to have become infected in India. One such strain that was derived from a patient who came to Japan from India was isolated, and a nearly complete HEV genome sequence has been reported for this isolate.27
One of the limitations of the present study is that sequence data were obtained only from a subset of the reported cases. This shortcoming posed several challenges. First, specimens were not obtained from patients in all reported cases. Second, viral sequences were obtained only from some of the collected sample because of nondetection of viral sequence, possibly due to virus levels being below detectable limits. Third, some municipalities do not collect sequence data, making it difficult to exclude the effect of regional bias. Nevertheless, we were able to obtain HEV sequences from 20% of the reported cases from 2018 to 2021, and the molecular findings, determined following sample collection, cannot be biased by the presumed place of infection reported by the case-patient. Our data permitted us to describe and speculate on national trends and distributions in HEV infection in Japan. Further cooperation from additional prefectural and municipal PHIs is expected to provide more reliable national data.
In conclusion, enhanced national surveillance and molecular characterization studies of HEV in Japan from 2016 to 2021 revealed that cases of hepatitis E typically reflected infection from domestic sources, with most patients consisting of males in their 40s–70s. The consumption of pork and the meat of wild boar, in order, were reported as being the most likely routes of infection in these reported domestic cases. The majority of domestic HEV strains belonged to Genotypes 3a and 3b. In contrast, most HEV G1 strains were detected in patients who appeared to have been infected abroad. Continuing efforts should be made to investigate the molecular epidemiology of HEV in animal reservoirs and in food. Such investigations are expected to clarify the routes of transmission of HEV in Japan, as well as shifts in circulating subtypes, as has been observed in other countries.28 The consumption of products containing raw or undercooked pig liver, blood, and meat products likely represents the major potential risk factor for human HEV infections in Japan. Adaptations of food production processes (e.g., heat treatment) might reduce hepatitis E infection.
AUTHOR CONTRIBUTIONS
Study concept and design: Ryuichi Sugiyama, Osamu Takahara, Tomoko Kiyohara, Koji Ishii, Ryosuke Suzuki. Acquisition of data and technical support: Ryuichi Sugiyama, Osamu Takahara, Yuichiro Yahata, Kazuhiko Kanou, Mami Nagashima, Tian-Cheng Li, Yuzo Arima, Hiroto Shinomiya, Koji Ishii, Ryosuke Suzuki. Analysis and interpretation of data: Ryuichi Sugiyama, Osamu Takahara, Yuichiro Yahata, Yuzo Arima, Masamichi Muramatsu, Ryosuke Suzuki. Drafting of the manuscript: Yuichiro Yahata, Yuzo Arima, Ryosuke Suzuki. Critical revision of the manuscript: All authors. Study supervision: Yuzo Arima, Koji Ishii, Masamichi Muramatsu, Ryosuke Suzuki.
ACKNOWLEDGMENTS
This research was based on cases submitted by prefectural and municipal PHIs and PHCs to the NESID system. Some of the cases were diagnosed based on microbiological tests performed at PHIs/PHCs. We sincerely appreciate the efforts of staff members from the PHIs and PHCs, along with the reporting physicians, for providing information on the infectious diseases included in this study. The following PHIs/PHCs cooperated with the molecular epidemiologic study: Otaru City Health Center, Research Institute for Environmental Sciences and Public Health of Iwate Prefecture, Morioka City Health Center, Sendai City Institute of Public Health, Miyagi Prefectural Institute of Public Health and Environment, Kurihara Health Center, Akita Research Center for Public Health and Environment, Fukushima Institute of Public Health, Ibaraki Prefectural Institute of Public Health, Mito City Health Center, Utsunomiya City Institute of Public Health and Environment, Gunma Prefectural Institute of Public Health and Environmental Sciences, Takasaki City Health Center, Maebashi City Health Center, Saitama Institute of Public Health, Kawaguchi City Health Center, Chiba City Institute of Health and Environment, Chiba Prefectural Institute of Public Health, Funabashi City Health Center, Shinjuku City Public Health Center, Edogawa Public Health Center, Tokyo Metropolitan Institute of Public Health, Kanagawa Prefectural Institute of Public Health, Fujisawa City Health Center, Kawasaki City Institute for Public Health, Yokohama City Institute of Health, Niigata Prefectural Institute of Public Health and Environmental Sciences, Toyama City Health Center, Ishikawa Prefectural Institute of Public Health and Environmental Science, Fukui Prefectural Institute of Public Health and Environmental Science, Yamanashi Institute for Public Health, Nagano Environmental Conservation Research Institute, Gifu Prefectural Research Institute for Health and Environmental Sciences, Gifu City Department of Health and Hygiene, Shizuoka City Institute of Environmental Sciences and Public Health, Shizuoka Institute of Environment and Hygiene, Hamamatsu Institute of Health and Environment, Mie Prefecture Health and Environment Research Institute, Shiga Prefectural Institute of Public Health, Otsu City Health Center, Kyoto City Institute of Health and Environmental Sciences, Osaka City Institute of Public Health and Environmental Sciences, Higashiosaka City Environmental Sanitation Inspection Center, Osaka Institute of Public Health, Public Health Research Institute of Kobe City, Himeji City Environment Sanitary and Hygiene Research Institute, Nishinomiya City Health Center, Kobe Institute of Health, Nara City Health Center, Nara Prefectural Institute for Hygiene and Environment, Tottori Prefecture Division of Health and Welfare, Tottori Prefectural Institute of Public Health and Environmental Science, Okayama City Health Center, Kurashiki City Health Center, Hiroshima City Institute of Public Health, Yamaguchi Prefectural Institute of Public Health and Environment, Takamatsu City Health Center, Fukuoka City Institute for Hygiene and Environment, Kitakyushu City Institute of Health and Environmental Sciences, Saga Prefectural Institute of Public Health and Pharmaceutical Research, Miyazaki Prefectural Institute for Public Health and Environment, Kagoshima City Health Center. The authors of this manuscript express their gratitude to Y. Hirama and M. Oizumi for administrative assistance. This research was supported in part by Grants-in-Aid from the Ministry of Health, Labour and Welfare (MHLW Grant No. 10KA1006), the Japan Society for the Promotion of Science of KAKENHI (JP23K07934), and from the Japan Agency for Medical Research and Development (AMED Grant Nos. JP22fk0108139, JP23fk0210109, and JP23fk0108683).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The case patient data are publicly available in Japanese: https://www.niid.go.jp/niid/ja/hepatitis-e-m/hepatitis-e-iasrtpc/10837-502t.html.