Clinical isolates of hepatitis B virus genotype C have higher in vitro transmission efficiency than genotype B isolates
Abstract
Serum samples were collected from 54 hepatitis B e antigen (HBeAg)-positive Chinese patients infected with hepatitis B virus (HBV) subgenotype B2 or C2. They were compared for transmission efficiency using same volume of samples or infectivity using same genome copy number. Adding polyethylene glycol (PEG) during inoculation did not increase infectivity of fresh samples but markedly increased infectivity following prolonged sample storage. Differentiated HepaRG cells infected without PEG produced more hepatitis B surface antigen (HBsAg) and higher HBsAg/HBeAg ratio than sodium taurocholate cotransporting polypeptide (NTCP)-reconstituted HepG2 cells infected with PEG. They better supported replication of core promoter mutant in contrast to wild-type (WT) virus by HepG2/NTCP cells. Overall, subgenotype C2 samples had higher viral load than B2 samples, and in general produced more HBeAg, HBsAg, and replicative DNA following same-volume inoculation. Precore mutant was more prevalent in subgenotype B2 and had reduced transmission efficiency. When same genome copy number of viral particles was inoculated, viral signals were not necessarily higher for three WT C2 isolates than four WT B2 isolates. Using viral particles generated from cloned HBV genome, three WT C2 isolates showed slightly reduced infectivity than three B2 isolates. In conclusion, subgenotype C2 serum samples had higher transmission efficiency than B2 isolates in association with higher viral load and lower prevalence of precore mutant, but not necessarily higher infectivity. PEG-independent infection by HBV viremic serum samples is probably attributed to a labile host factor.
Abbreviations
-
- ALT
-
- alanine transaminase
-
- BCP
-
- basic core promoter
-
- cHBV
-
- cell culture-derived HBV
-
- ELISA
-
- enzyme-linked immunosorbent assay
-
- FBS
-
- fetal bovine serum
-
- HBeAg
-
- hepatitis B e antigen
-
- HBsAg
-
- hepatitis B surface antigen
-
- HBV
-
- hepatitis B virus
-
- HCC
-
- hepatocellular carcinoma
-
- IC
-
- immune clearance
-
- IP
-
- immunoprecipitation
-
- IT
-
- immune tolerance
-
- NTCP
-
- sodium taurocholate cotransporting polypeptide
-
- PC
-
- precore
-
- PCR
-
- polymerase chain reaction
-
- PEG
-
- polyethylene glycol
-
- PHH
-
- primary human hepatocyte
-
- qPCR
-
- quantitative PCR
-
- sHBV
-
- serum-derived HBV
-
- SVP
-
- subviral particle
-
- WT
-
- wild-type
1 INTRODUCTION
Hepatitis B virus (HBV) infection can be transmitted by blood transfusion, shared needles, sex, and also vertically from infected mothers. Hepatitis B surface antigen (HBsAg) and hepatitis B e antigen (HBeAg) serve as sensitive serological markers of ongoing HBV infection. Most adulthood transmission leads to self-limited acute infection, with HBsAg positivity lasting for <6 months. In contrast, over 90% of perinatal transmission from HBeAg+ mothers to the offspring ends up as chronic infection lasting for several decades, which greatly increases the risk to develop hepatocellular carcinoma (HCC).1-3 HBeAg and overproduced HBsAg are implicated in the induction of immune tolerance (IT) to promote chronic HBV infection.4 HBeAg is a secreted variant of core protein,5 the building block of core particles that support HBV genome replication. HBsAg corresponds to viral envelope proteins on the 42-nm virions and 22-nm subviral particles (SVPs) lacking internal core particles. Such noninfectious SVPs exceed virions by >1000 fold in patient blood.6, 7 Since IT induction is most efficient at a younger age, chronic HBV infection is most common in East Asia in association with perinatal transmission of genotype B or C. The IT phase of chronic HBV infection is characterized by positive HBeAg and HBsAg, high viral load, and a homogeneous population of wild-type (WT) HBV sequence. Break of IT will lead to reduced viremia, with sequential HBeAg and HBsAg seroconversion (loss of the antigen followed by rise of corresponding antibody). The early immune clearance (IC) phase may select for G1896A precore (PC) mutation to abolish HBeAg at the translational level,8-11 or A1762T/G1764A basic core promoter (BCP) mutations to reduce HBeAg expression while enhancing genome replication at the transcript level.12-16
Although genotypes B and C have overlapping geographic distributions, shared transmission routes and target populations, genotype C patients experience a decade delay in HBeAg seroconversion.17-19 That increases their lifelong HCC risk.20-23 The biological bases for different clinical features of the two HBV genotypes remain poorly understood. We previously cloned HBV genome from serum samples of Chinese patients infected with genotype B (subgenotype B2) or C (subgenotype C2), and transfected dimeric HBV DNA construct into Huh7 human hepatoma cell line. The WT (in terms of BCP and PC sequences) C2 isolates displayed lower replication capacity than WT B2 isolates due to a weaker core promoter.24, 25 They had more efficient virion secretion as evidenced by a higher ratio of extracellular virion DNA/intracellular replicative DNA.24 Whether the two genotypes differ at the earlier step of viral entry into hepatocytes has not been systemically studied. In this regard, HBV can infect differentiated HepaRG cells or HepG2 cells reconstituted with sodium taurocholate cotransporting polypeptide (NTCP), the functional HBV receptor.26-28 Here we employed these two cell lines for comparative infection experiments of B2 and C2 clinical isolates. Initially, same volume of patient serum samples was inoculated to compare their relative transmission efficiency. Next same genome copy number was used to compare infectivity of a few WT isolates. Finally HBV genome was cloned from such serum samples, and viral particles secreted from cells transfected with the replication construct were compared for infectivity.
2 MATERIALS AND METHODS
2.1 Serum samples, genotyping, and check for mutations
Serum samples were collected from 54 HBeAg+ and treatment-naïve Chinese patients infected with subgenotype B2 or C2. Serum HBV DNA, HBsAg, and HBeAg were quantified by ADICON Clinical Laboratories using COBAS@ TaqMan@ platform and Abbott ARCHITECT i2000 platform, respectively. Liver biopsy or dynamic serum alanine transaminase (ALT) and HBV DNA levels within 6 months of follow up were available for 24 patients to classify them into the IT or IC phase of chronic infection. The 3.2-kb HBV genome was amplified from serum samples by polymerase chain reaction (PCR) and sequenced directly. Viral genotype was determined by phylogenetic analysis. BCP and PC mutants were defined by presence of A1762T/G1764A or G1896A mutation.
2.2 In vitro HBV infection using viremic serum samples
The HepG2 cell line stably transfected with an NTCP expression construct (HepG2/NTCP) and HepaRG cell line have been described.29, 30 Cells seeded in 96-well plates were used for infection, with confluent HepaRG cells differentiated for >2 weeks before infection. To compare in vitro transmission efficiency, 2 μL of serum sample was incubated with cells for 16 h in medium not supplemented with fetal bovine serum (FBS), with 4% polyethylene glycol (PEG) for HepG2/NTCP cells but without PEG for HepaRG cells. Cells were washed and further cultured in FBS-containing medium. HBsAg and HBeAg were measured from culture supernatant through enzyme-linked immunosorbent assay (ELISA) using commercial kits (Kehua), using a volume not causing signal saturation. Cells were harvested at Day 20 postinfection for Southern blot using a mixed probe of B2 and C2 subgenotypes.30 To compare HBV infectivity using same genome copy number, serum samples were diluted with normal human serum.
2.3 1.1mer construct and production of cHBV particles
Generation of 1.1mer construct from serum samples has been described,30 with a 14-nt distance between the end of CMV promoter and predicted transcription initiation site for pregenomic RNA. The 3.2-kb HBV genome was amplified by PF1'—PR1 primer pair, and an overlapping 0.1-kb fragment using PF2—PR2 primer pair. The two DNA fragments were cloned to pcDNA3.1zeo(-) vector using a DNA assembly kit. HepG2 cells in six-well plates were transfected with 2 μg of 1.1mer construct using TransIT-LT1 reagent (Mirus). Cell culture-derived HBV (cHBV) particles from combined Days 3 and 5 culture supernatant were PEG precipitated.29 They were resuspended in 1/30th−1/70th original volume, and inoculated overnight with 4% PEG but no FBS, into HepG2/NTCP or HepaRG cells seeded in 96-well plates. To inoculate with same genome copy number of viral particles, virion DNA was quantified by PCR from a fraction of culture supernatant following immunoprecipitation with anti-preS1 and anti-S antibodies.30
2.4 Analysis of HBV markers from transfected cells
HBV RNAs and replicative DNA from HepG2 cells transfected with the 1.1mer construct were detected by Northern and Southern blot with 32P-labled full-length probe, while envelope and core proteins were detected by Western blot.16 Virions were immunoprecipitated from culture supernatant followed by PCR quantification of HBV DNA. Alternatively, virions and core particles were separated through native agarose gel electrophoresis followed by transfer to nitrocellulose filter and hybridization with HBV DNA probe.
2.5 Statistical analysis
Differences between the groups were examined using a Student t test or Wilcoxon Ranksum test by GraphPad Prism 6 (GraphPad Software, Inc.). A p value of <0.05 is considered as statistically significant. Data are presented as means ± standard deviations (SD).
3 RESULTS
3.1 Serum samples for functional characterization
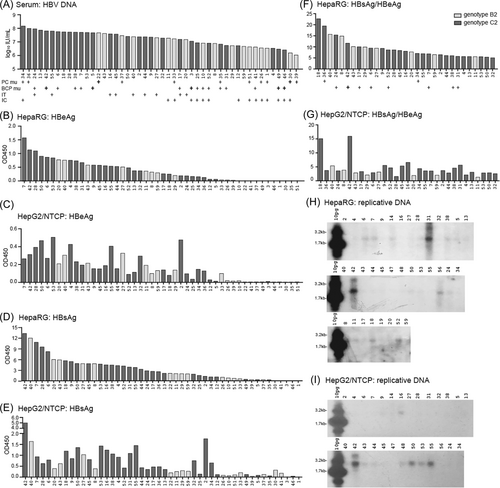
Serum samples were collected from 54 HBeAg+ and treatment-naïve Chinese patients. Most data analysis was based on 48 samples with information on viral genotype, BCP and PC sequences: 30 of C2 and 18 of B2 subgenotypes (Table 1). They had HBV DNA titer of 1.07 × 106−1.44 × 108 IU/mL (Figure 1A). The G1896A PC mutation and A1762T/G1764A BCP mutations were found in 11 and 16 samples, respectively (Figure 1A; sample #35 had both PC and BCP mutations), as a pure population or mixture with WT sequence (Table 2). Consistent with the higher prevalence of PC mutation in genotype B but BCP mutations in genotype C,19, 31-33 8 of 10 samples with just PC mutation belonged to B2 subgenotype, whereas 13 of the 15 samples with BCP mutations only were of C2 subgenotype (Table 2).
| Genotype B2: 18 | Genotype C2: 30 | p Value | |
|---|---|---|---|
| Male gender | 10 (55.6%) | 21 (70.0%) | 0.311 |
| Age (year) | 28 (25–29) | 34 (30–38) | 0.003 |
| ALT (U/L) | 86 (42–266) | 54 (31–86) | 0.078 |
| HBV DNA (log10 IU/mL) | 6.90 (6.60–7.33) | 7.49 (6.92–7.74) | 0.004 |
| HBeAg (PEI-U/mL) | 1975 (1138–2663) | 3600 (2300–4613) | 0.003 |
| HBsAg (log10 IU/mL) | 4.53 (4.17–4.83) | 4.69 (4.40–4.80) | 0.509 |
| HBeAg: HepaRG | 0.038 (0.010–0.397) | 0.426 (0.146–0.737) | 0.024 |
| HBsAg: HepaRG | 1.452 (0.331–4.786) | 3.449 (1.180–4.798) | 0.120 |
| HBeAg: HepG2/NTCP | 0.060 (0.007–0.196) | 0.153 (0.068–0.300) | 0.043 |
| HBsAg: HepG2/NTCP | 0.228 (0.116–0.384) | 0.699 (0.294–1.116) | 0.013 |
| Infectious in HepaRG | 16 (88.9%) | 28 (93.3%) | 0.590 |
| Infectious in HepG2/NTCP | 15 (83.3%) | 26 (86.7%) | 0.751 |
- Abbreviations: ALT, alanine transaminase; HBeAg, hepatitis B e antigen; HBsAg, hepatitis B surface antigen.
- a Serum samples (2 μL) were inoculated to HepG2/NTCP or HepaRG cells seeded in 96-well plates, followed by measurement of HBeAg and HBsAg titers 15 days later. Shown are values from 50 μL of culture supernatant (OD450). Quantitative variables are presented as median followed by interquartile ranges.
| WT: 22(B2: 8; C2: 14) | BCP mutant: 15b (B2: 2; C2: 13) | PC mutant: 10c (B2: 8; C2: 2) | p Value | |
|---|---|---|---|---|
| Male gender | 14 (63.6%) | 12 (80.0%) | 4 (40%) | 0.125 |
| Age (year) | 31 (26–35) | 32 (27–37) | 28 (27–37) | 0.774 |
| ALT (U/L) | 34 (27–58) | 79 (52–123) | 102 (58–295) | 0.002 |
| HBV DNA (log10 IU/mL) | 7.37 (7.10–7.64) | 7.05 (6.74–7.71) | 6.94 (6.45–7.75) | 0.550 |
| HBeAg (PEI-U/mL) | 3150 (2463–4238) | 3100 (1050–4550) | 1328 (388–2663) | 0.032 |
| HBsAg (log10 IU/mL) | 4.72 (4.59–4.87) | 4.54 (3.70–4.79) | 4.35 (3.82–4.54) | 0.008 |
| HBeAg: HepaRG | 0.532 (0.316–0.755) | 0.181 (0.010–0.732) | 0.019 (0.000–0.062) | <0.001 |
| HBsAg: HepaRG | 4.543 (1.996–5.437) | 2.610 (0.328–4.383) | 0.593 (0.272–1.308) | 0.001 |
| HBeAg: HepG2/NTCP | 0.201 (0.138–0.396) | 0.090 (0.011–0.232) | 0.011 (0.006–0.106) | <0.001 |
| HBsAg: HepG2/NTCP | 0.778 (0.373–1.269) | 0.348 (0.083–0.933) | 0.197 (0.100–0.359) | 0.003 |
- Abbreviations: ALT, alanine transaminase; BCP, basic core promoter; HBeAg, hepatitis B e antigen; HBsAg, hepatitis B surface antigen; HBV, hepatitis B virus; PC, precore; WT, wild-type.
- a Serum samples (2 μL) were inoculated to HepG2/NTCP or HepaRG cells seeded in 96-well plates, followed by measurement of HBeAg and HBsAg titers 15 days later.
- b Five of the 15 samples harbored pure A1762T/G1764A BCP mutations based on Sanger sequencing (#3, #5, #42, #46, #49), all belonging to C2 subgenotype. The others (#6, #7, #10, #17, #24, #25, #31, #38, #41, #52) had mixture with WT HBV.
- c Two of the 10 samples harbored pure G1896A mutation based on Sanger sequencing (#22, #30), both belonging to B2 subgenotype. The others (#1, #26, #33, #34, #36, #37, #39, #51) had mixture with WT HBV. Shown are values from 50 μL of culture supernatant (OD450). Excluded from analysis are five serum samples with indeterminate BCP or PC sequence, as well as sample #35 with both BCP and PC mutations. Quantitative variables are presented as median followed by interquartile ranges.
3.2 sHBV infected HepaRG cells better supported replication of BCP mutant in contrast to WT virus by HepG2/NTCP cells
To identify highly infectious HBV isolates and to compare transmission efficiency we first inoculated 2 μL of serum samples into HepG2/NTCP cells or differentiated HepaRG cells seeded in 96-well plates. Considering PEG-independent infection of HepaRG cells by serum-derived HBV (sHBV) particles,29 inoculation was performed without PEG for HepaRG cells but with 4% PEG for HepG2/NTCP cells. Forty-four and 41 of the 48 samples were respectively infectious in HepaRG cells and HepG2/NTCP cells (Table 1), as defined by positive HBeAg and/or HBsAg from culture supernatant at Day 15 postinoculation. Thirty-nine samples were infectious in both cell types, five in HepaRG cells only, and two marginally infectious in HepG2/NTCP cells only. Consistent with our previous report,29 HepaRG cells produced much higher HBsAg titer than HepG2/NTCP cells (Table 1; Figure 1B–E) and higher HBsAg/HBeAg ratio for most samples (Figure 1F vs. 1G; Supporting Information: Table S1). That was also true when comparison was made within WT isolates or among BCP mutants (Supporting Information: Table S2). In HepaRG cells sample #42 and #31 produced highest level of replicative DNA, followed sequentially by #6, #7, #14, #56, #38, #52, #18, #24, and #11 (Figure 1H). Most such samples harbored BCP mutations, with only #11 and #18 having pure WT sequence (Samples #14 and #56 had no clear sequencing data). In HepG2/NTCP cells level of replicative DNA was #42 > #55 > #50, with weak signals from #16, #53, #43, #44, and #48 (Figure 1I). They had pure WT sequence except #42, which had BCP mutations (Sample #48 had no clear sequencing data).
3.3 C2 samples had higher HBV DNA titers than B2 samples and higher transmission efficiency
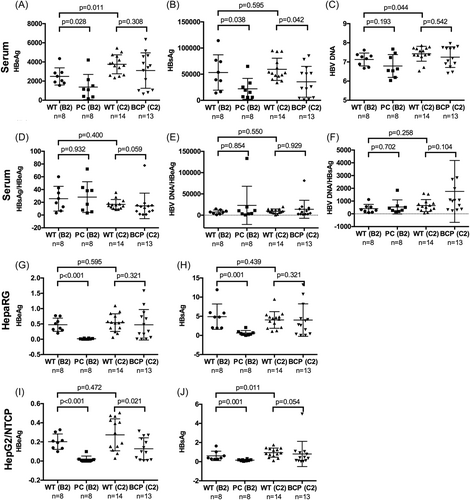
The 30 C2 samples had higher median and mean titers of HBV DNA and HBeAg than the 18 B2 samples (Table 1; Supporting Information: Figure S1A,C). Slightly higher percentage of C2 samples was infectious than B2 samples in both cell types (Table 1). The C2 samples also produced higher titers of HBeAg and HBsAg than B2 samples, although for HepaRG cells the difference in HBsAg did not reach statistical significance (Table 1; Supporting Information: Figure S1D–G). In HepaRG cells 9 of the 11 isolates with high replication were of C2 subgenotype. Only #11 was a B2 isolate while the genotype for #14 was undetermined. Of the eight isolates with high DNA replication in HepG2/NTCP cells, all but #43 and #48 were C2 isolates. When only samples with WT BCP and PC sequences were compared, the 14 C2 serum samples had higher mean HBeAg and HBV DNA titers than the eight WT B2 samples (Figure 2A,C). They produced higher mean HBeAg and HBsAg titers than the B2 samples in HepG2/NTCP cells, with the difference in HBeAg not statistically significant (Figure 2I,J). In this cell line five WT C2 samples (#16, #44, #50, #53, #55) but only one WT B2 sample (#43) showed high replication (Figure 1I).
3.4 Reduced transmission efficiency of PC mutant than WT HBV
Serum samples of the eight PC mutants of B2 subgenotype had lower mean HBeAg, HBsAg, and HBV DNA titers than those of the eight WT B2 samples, although the difference in HBV DNA titer did not reach statistical significance (Figure 2A–C, left). Since in six of the eight samples (#1, #26, #33, #37, #39, #51) the PC mutant co-existed with WT virus, their median HBsAg/HBeAg ratio and HBV DNA/HBeAg ratio were not much elevated (Figure 2D,E). They produced lower HBsAg titer in both HepaRG and HepG2/NTCP cells (Figure 2H,J) suggesting reduced transmission efficiency. Surprisingly the two C2 samples harboring just PC mutation (#34, #36) had the highest HBV DNA titers among the 48 samples (Figure 1A), but they produced low HBeAg and HBsAg titers in HepG2/NTCP and especially HepaRG cells (Figure 1B–E).
Compared with the 14 WT C2 samples, the 13 C2 samples with BCP mutations but no CP mutation had lower mean HBeAg, HBsAg, and HBV DNA titers, with the difference in HBsAg titer being statistically significant (Figure 2A–C). These samples produced less HBeAg and HBsAg than the WT C2 samples in HepG2/NTCP cells, but no statistically significant difference was observed in HepaRG cells (Figure 2G–J). While all but #42 of the six high replicating C2 isolates in HepG2/NTCP cells had WT sequence, seven of the eight C2 isolates with high replication in HepaRG cells were BCP mutant (only #18 had WT sequence).
One concern with the above analysis was the small sample size, making it difficult to reach statistically significant findings. We, therefore, compared WT virus, BCP mutant, and PC mutant irrespective of viral genotype. Serum HBV DNA, HBsAg, and HBeAg titers were highest for the 22 WT isolates, intermediate for the 15 BCP mutants, and lowest for the 10 PC mutants, although the difference in HBV DNA titer did not reach statistical significance (Table 2). The median ALT level was highest for PC mutants and lowest for WT isolates. Median HBsAg and HBeAg titers in infected HepaRG and HepG2/NTCP cells were also highest for WT isolates and lowest for the PC mutants (Table 2). Consistent with reduced HBeAg expression by BCP mutations, such samples produced higher HBsAg/HBeAg ratio than WT isolates in HepaRG and especially HepG2/NTCP cells (Supporting Information: Table S2).
3.5 PEG could enhance sHBV infectivity, especially following prolonged sample storage
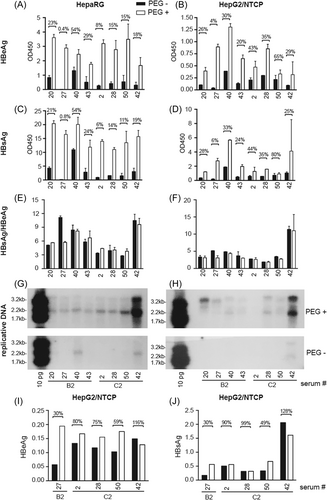
While data presented above revealed higher transmission efficiency of C2 than B2 isolates, and WT virus than PC mutant when same volume of serum sample was inoculated, it remained uncertain whether WT C2 isolates had higher infectivity than WT B2 isolates when same number of viral particles was inoculated. Also, to compare the two cell lines for susceptibility to HBV infection required identical inoculation condition (with or without PEG). We selected four B2 isolates and four C2 isolates for such comparison. All had high HBV DNA titers (1.48–4.18 × 107 genomes/μL) with infectivity in both cell lines when 2 μL of serum sample was inoculated. All harbored WT BCP and PC sequences except for #42, which had BCP mutant as a pure population. This sample was included because it produced highest HBsAg and replicative DNA in both cell types (Figure 1D,E,H,I), with higher HBsAg/HBeAg ratio in HepG2/NTCP than HepaRG cells in contrast to most other samples (Figure 1F,G). Cells in 96-well plates were inoculated with serum samples diluted with normal human serum to reach 10 genomes/cell. When inoculated with 4% PEG, differentiated HepaRG cells produced higher titers of HBeAg and especially HBsAg than HepG2/NTCP cells, most striking for #20, #2, and #50 (Figure 3A–D). HepaRG cells also produced higher HBsAg/HBeAg ratio with the notable exception of #42 (Figure 3E,F), although the higher ratio in HepaRG cells was more striking in the initial screening (Table 3). Inoculation without PEG reduced HBV infectivity for both cell types whether according to HBeAg, HBsAg, or HBV DNA (Figure 3A–D,G,H). The PEG dependence was highest for #27, and least for #40 in HepaRG cells but #50 in HepG2/NTCP cells.
| Serum sampleb | Genotype | HepaRG no PEG (Figure 1F) | HepaRG + PEG (Figure 3E) | HepaRG + PEG (Figure 5K) | HepG2/NTCP + PEG (Figure 1G) | HepG2/NTCP + PEG (Figure 3F) | HepG2/NTCP + PEG (Figure 5G) |
|---|---|---|---|---|---|---|---|
| #2 | C2 | 6.46 | 4.37 | 4.12 | 3.70 | 2.63 | 2.78 |
| #20 | B2 | 7.73 | 5.65 | 7.84 | 1.89 | 3.13 | 1.58 |
| #27 | B2 | 9.41 | 5.70 | 10.36 | 3.15 | 3.11 | 1.76 |
| #28 | C2 | 8.64 | 4.05 | 3.85 | 1.57 | 1.87 | 1.07 |
| #40 | B2 | 15.70 | 8.18 | N.A. | 5.38 | 4.35 | N.A. |
| #42 | C2 | 11.65 | 9.61 | 17.71c | 15.92 | 11.03 | 14.92c |
| #43 | B2 | 10.09 | 6.69 | 10.27 | 2.85 | 3.03 | 1.89 |
| #50 | C2 | 5.36 | 3.79 | ND | 2.90 | 2.76 | ND |
| #55 | C2 | 6.92 | N.A. | 6.36 | 3.57 | N.A. | 1.59 |
- Abbreviations: BCP, basic core promoter; cHBV, cell culture-derived HBV; HBeAg, hepatitis B e antigen; HBsAg, hepatitis B surface antigen; HBV, hepatitis B virus; PC, precore; PEG, polyethylene glycol; sHBV, serum-derived HBV.
- a Values in Figure 1F,G were from relatively fresh serum samples, while values in Figure 3E,F were from samples with prolonged storage. Values from Figure 5K,G were from cloned genome (as cHBV rather than sHBV).
- b All the serum samples had WT BCP and PC sequence except for #42, which had 1762/1764 BCP mutations as a pure population in addition to 5' preS1 deletions as quasispecies.
- c Not from Figure 5K or Figure 5G but from separate experiments. Based on clone 42.5, which had intact genome of 3215nt. Clones 42.1 and 42.7 from the same serum sample harbored 15-nt or18-nt deletion at 5' preS1 deletion to shorten L protein by 11aa. All the three clones contained 1762/1764 BCP mutations.
The PEG dependence of sHBV infectivity in HepaRG cells contradicted our previous finding,29 for which viral particles concentrated from relatively fresh serum samples by PEG precipitation or ultracentrifugation through sucrose gradient were inoculated. Data presented in Figure 1 and Figure 3A–H were generated 3–11 months and 16–24 months after sample collection, respectively. Another infection experiment of some of these samples in HepG2/NTCP cells 12 months earlier than Figure 3B,D,F manifested much less PEG dependence, whether according to HBeAg or HBsAg (Figure 3I,J).
3.6 No evidence for higher infectivity of serum samples of three WT C2 isolates than four WT B2 isolates
Among the eight serum samples selected to compare infectivity, #42 had the highest HBV DNA titer. With same genome copy number for inoculation and in the presence of PEG, it no longer produced highest HBsAg titer but rather low HBeAg titer (Figure 3A–D). It produced higher HBsAg/HBeAg ratio than other samples, especially in HepG2/NTCP cells (Figure 3E,F). It also produced much higher replicative DNA than the other seven samples, especially in HepaRG cells (Figure 3G,H, top panels). In this regard, #42 had 1762/1764 BCP mutations as a pure population in contrast to WT sequence of the other seven samples, and such mutations augment HBV genome replication at the transcript level.13-16, 24 Comparison among the seven WT samples inoculated with PEG did not reveal higher infectivity of the C2 isolates. First, there was isolate-to-isolate variability within the same subgenotype. Second, findings from the two cell types were not always concordant. Third, ranking based on HBsAg did not always agree with that according to HBeAg. Indeed the seven samples differed in their HBsAg/HBeAg ratios (Figure 3E,F), and ranking from replicative DNA could also differ.
3.7 Three WT C2 clones as 1.1mer construct generated more replicative DNA and virions than three WT B2 clones in transfected HepG2 cells
To avoid complications from viral quasispecies (coexistence of BCP or PC mutant with WT virus), or possible host factors modulating HBV infectivity, we cloned HBV genome from viremic serum samples to generate cHBV particles. In this regard, a 1.1mer construct is often used to maximize pgRNA transcription, genome replication, and virion production.34-36 For the eight serum samples compared for relative infectivity (Figure 3), three WT B2 isolates (#20, #27, #43) and two WT C2 isolates (#2, #28) were used to generate 1.1mer construct. Another WT C2 isolate for cloning was #55 with high HBV DNA titer and high transmission efficiency in HepG2/NTCP cells (Figure 1). For each isolate a 1.1mer clone with its HBV DNA sequence identical to the consensus sequence of the original serum sample was chosen. In transiently transfected HepG2 cells the 1.1mer construct of the three WT C2 clones in general produced higher level of core protein and replicative DNA than the three WT B2 clones (Figure 4B,C). They secreted more virions and naked core particles according to native agarose gel electrophoresis (Figure 4D). More virion secretion by the C2 clones was confirmed by immunoprecipitation followed by qPCR (Figure 4E).
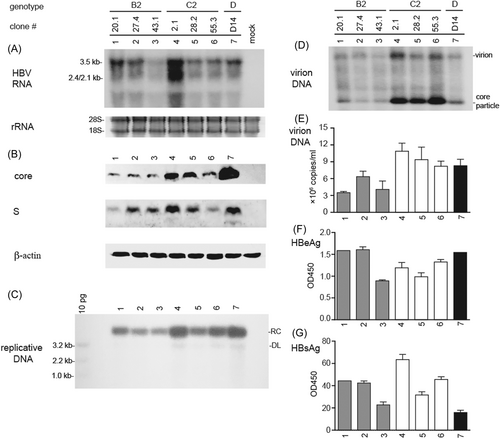
3.8 The three WT C2 clones had slightly reduced infectivity than the three WT B2 clones
cHBV particles concentrated from culture supernatant of transfected HepG2 cells were inoculated to HepG2/NTCP cells or differentiated HepaRG cells in the presence of 4% PEG. When same genome copy number of viral particles was used, B2 clone 27.4 produced somewhat higher level of HBeAg, HBsAg, and replicative DNA than the two other B2 clones in HepG2/NTCP cells (Figure 5E,F,H). For the three C2 clones the three parameters did not agree with each other. Overall viral signals from C2 clones appeared reduced than B2 clones in this cell line, especially for replicative DNA (Figure 5E,F,H). Consistent with more efficient virion production by the C2 clones as 1.1mer construct in transfected HepG2 cells (Figure 4D,E), inoculation with viral particles concentrated from same volume of culture supernatant led to increased signals for the C2 than B2 clones (compare Figure 5A,B,D with Figure 5E,F,H). In HepaRG cells inoculated with same genome copy number of viral particles, the C2 clones produced lower HBsAg titer and HBsAg/HBeAg ratio, and less replicative DNA than the B2 clones (Figure 5I–L).

3.9 Impact of sHBV-to-cHBV conversion on HBsAg/HBeAg ratio
HepaRG cells produced higher HBsAg/HBeAg ratio than HepG2/NTCP cells, most striking for the genotype D clone serving as a positive control (compare Figure 5C,G,K). After molecular cloning to convert sHBV particles into cHBV particles, the HBsAg/HBeAg ratio in HepaRG cells inoculated with PEG remained similar for sample #20, #27, #43, and #55 but reduced for #2 and especially #28 when compared with data from Figure 1F (fresh serum samples inoculated without PEG) (Table 3). But if comparison was made with stored serum samples inoculated also with PEG (Figure 3E), the ratio was rather increased for #20, #27, #43. In this regard, the HBsAg/HBeAg ratio was reduced in Figure 3E than in Figure 1F for all the samples tested (Table 3). For HepG2/NTCP cells the ratio decreased for most samples. The much higher HBsAg/HBeAg ratio in HepaRG cells than HepG2/NTCP cells sustained or became even more striking (compare Figure 5G with Figure 5K).
4 DISCUSSION
HBV genotypes B and C are responsible for majority of chronic HBV infection in East Asia, with B2 and C2 being the most common in most parts of China. A major aim of the present study was to compare their transmission efficiency by inoculating same volume of serum samples. This has relevance to spread of HBV infection where a small volume of blood or body fluid was involved through shared needles, mosquito bites, sex, visits to barber's shop, or maternal-infantile transmission. Consistent with prolonged HBeAg+ phase of genotype C than genotype B infection,17-19 the 30 C2 patients were older and had higher HBeAg and HBV DNA titers but lower ALT levels than the 18 B2 patients (Table 1; Supporting Information: Figure S1A,C). Of the 24 patients with liver biopsy or serial serum samples, 6 of 13 C2 patients but only 3 of 11 B2 patients were classified into the IT phase (Figure 1A). In both HepG2/NTCP and HepaRG cells inoculated with 2 μL of serum samples, the C2 isolates produced higher HBsAg and HBeAg titers than the B2 samples (Table 1; Supporting Information: Figure S1D–G), with most high replicating samples belonging to C2 subgenotype (Figure 1H,I). These findings suggested higher transmission efficiency of C2 isolates, which could be at least partly attributed to lower prevalence of PC mutant in C2 (3 of 30 samples) than B2 subgenotype (8 of 18 samples). The eight B2 samples with just PC mutation had lower HBV markers than the eight B2 samples with pure WT sequence (Figure 2A–C), and produced much lower mean HBeAg and HBsAg titers in both cell types (Figure 2G–J). Furthermore, none of the high-replicating samples in either cell type harbored PC mutation (Figure 1H,I). When WT virus, PC mutant, and BCP mutant were compared irrespective of viral genotype, HBsAg and HBV DNA titers in serum samples were highest for the WT isolates and lowest for the PC mutant (Table 2). Surprisingly, two C2 samples with just PC mutation (#34 and #36) had poor transmission efficiency in both cell lines (Figure 1D–G) despite having the highest HBV DNA titers among all the 48 samples (Figure 1A). HBeAg expression is dispensable for HBV genome replication according to in vitro transfection experiments,37, 38 and the G1896A PC mutation sustained infectivity of cHBV particles of a genotype D clone when inoculated to primary human hepatocytes (PHHs).39 Thus although HBeAg expression is critical for the induction of IT for the establishment of chronic HBV infection in vivo,4 it is most likely not required for HBV infection in cell culture. Whether the poor infectivity of serum samples #34 and #36 in vitro was attributed to other mutations in the HBV genome or host-derived antiviral factors remains an open question.
Whether BCP mutant also has reduced transmission efficiency than WT virus was less conclusive. Most BCP mutant (14 of 16 samples) belonged to C2 subgenotype. The 13 C2 samples harboring just BCP mutation had lower titers of HBV markers than the 14 C2 samples with pure WT sequence, with difference in HBsAg titer being statistically significant (Figure 2A–C). They produced lower mean HBsAg and HBeAg titers in HepG2/NTCP cells but not HepaRG cells (Figure 2G–J). All high replicating C2 isolates in HepG2/NTCP cells had WT sequence except for #42. For HepaRG cells only one of eight high-replicating C2 isolates had pure WT sequence. The remaining seven C2 isolates had BCP mutations coexisting with WT sequence or as a pure population (#42). One possible explanation for inconsistent findings from the two cell types is that differentiated HepaRG cells better support replication of BCP mutant than HepG2/NTCP cells. Indeed when inoculated with same genome copy number higher replication of #42 (a pure BCP mutant) than WT HBV isolates was more pronounced in HepaRG cells than HepG2/NTCP cells (Figure 3G vs. 3H). Alternatively or additionally, progeny HBV particles from infected HepaRG cells (but not HepG2/NTCP cells) can spread to neighboring cells. In this regard, the A1762T/G1764A BCP mutations increase genome replication,14-16, 24 which should augment virion production. Indeed a recent report found the ability of G1764A/C1766T BCP mutations (not the common A1762T/G1764A mutations) to markedly enhance infectivity of a genotype A isolate in PHHs co-cultured with human fibroblasts,40 apparently by more efficient spreading to adjacent hepatocytes because such mutations should not influence viral entry.
We also attempted to compare infectivity between B2 and C2 isolates by inoculating with same genome copy number of viral particles. To avoid the impact of BCP and PC mutations and viral quasispecies, we focused on seven pure WT isolates with high viral load and proven infectivity. Two of the four B2 isolates (#20 and #27) were known to be at the IT phase. There was no clear evidence for higher infectivity of the three WT C2 isolates than the four WT B2 isolates due to different rankings according to HBsAg, HBeAg, or replicative DNA, variable infectivity among samples of the same subgenotype, and discordant findings from the two cell lines (Figure 3). Overall the C2 samples produced lower HBsAg/HBeAg ratio than the B2 samples in both cell types. Following cloning of the HBV genome, the three WT C2 clones on average produced even slightly lower HBsAg titers and replicative DNA than the three WT B2 clones in both cell types when same genome copy number of viral particles was inoculated (Figure 5E–L). But following transient transfection of HepG2 cells with 1.1mer construct, the three C2 clones in general showed higher levels of genome replication, core protein expression, and virion production (Figure 4B–E). One way to better interpret the HBeAg, HBsAg, and replicative DNA data from infection experiment is to perform parallel transfection experiment in the same cell type (HepG2/NTCP cells) using dimeric HBV DNA construct. The 1.1mer construct prevents HBeAg expression, distorts the efficiency of genome replication, and might modify HBsAg secretion. Alternatively, considering the impacts of viral genotype, BCP and PC mutations, and other sequence differences on HBsAg versus HBeAg expression versus genome replication, covalently closed circular (ccc) DNA should serve as a more reliable marker of viral entry. cccDNA is the immediate product of incoming virion DNA and the template for viral RNA transcription, which precedes antigen production and genome replication.41, 42
Data interpretation for in vitro HBV infection is complicated by the inoculum (cHBV vs. sHBV) and target cells used (HepG2/NTCP, HepaRG, PHHs),43 and conditions of inoculation (with or without PEG). Most cHBV infections have employed viral particles concentrated from culture supernatant of HepG2 cells stably transfected with a single genotype D clone.35, 36, 44, 45 Using that genotype D clone, we found HepaRG cells maintained in 2% dimethyl sulfoxide (DMSO) produced lower HBeAg but higher HBsAg titer than similarly infected HepG2/NTCP cells cultured without DMSO.29 Others found 100-fold higher HBsAg/HBeAg ratio from HepaRG cells when HepG2/NTCP cells were also cultured with DMSO28! This could be attributed to overexpressed NTCP, which may up regulate the 3.5-kb RNA for HBeAg expression and genome replication.29 In the present study we found more striking cell type difference in the ratio for that genotype D clone than six clones of genotype B or C (Figure 5C,G,K). We also found higher HBsAg/HBeAg ratio from sHBV-infected HepaRG cells than HepG2/NTCP cells (Figure 1F,G). Although in that infection experiment, PEG was added for HepG2/NTCP cells but not HepaRG cells, later experiments found that PEG did not significantly alter the HBsAg/HBeAg ratio for either cell type (Figure 3E,F). Interestingly, the much higher HBsAg/HBeAg ratio in differentiated HepaRG cells than HepG2/NTCP cells for WT HBV could be reproduced after converting sHBV into cHBV (Table 3). Thus HepG2/NTCP cells better support HBeAg expression and genome replication (with preference for WT virus than BCP mutant) in contrast to more efficient HBsAg production by differentiated HepaRG cells, whether following cHBV or sHBV inoculation. We suspect that HepaRG cells, without NTCP overexpression, better mimic in vivo HBV infection in terms of HBsAg/HBeAg ratio. Indeed PHHs behaved like HepaRG rather than HepG2/NTCP cells in terms of that ratio following infection with cHBV of that genotype D clone (our unpublished observation). Large excess of SVP over virion production is a hallmark of in vivo HBV infection,6, 7 which renders HBsAg an unreliable marker of sHBV but not cHBV infectivity at early time points postinoculation.29
Efficient cHBV infection requires addition of 4% PEG during inoculation, whether for PHHs, HepaRG, or HepG2/NTCP cells.26-30, 43 PEG reportedly enhances virus attachment to target cells.46 We found partial virus purification by PEG precipitation or ultracentrifugation through sucrose cushion markedly enhanced infectivity of a few patient serum samples of genotype C.29 Such partially purified sHBV particles were poorly infectious in HepG2/NTCP cells, and their infectivity in HepaRG cells was not enhanced by the addition of 4% PEG during inoculation.29 The present study employed crude serum samples and omitted FBS during inoculation to better mimic natural HBV infection. In the presence of 4% PEG certain serum samples (notably #42, #50, #53, #55) could clearly infect HepG2/NTCP cells whether according to HBeAg, HBsAg, or replicative DNA. However, for most samples HepaRG cells produced higher HBeAg titer and much higher HBsAg titer than HepG2/NTCP cells despite inoculation without PEG (Figure 1B–E). They did not produce much higher level of replicative DNA, although samples of high replication did not overlap between the two cell lines except for #42 (Figure 1H,I). At a later time point, when both cell types were infected with serum samples in the presence of PEG, HepaRG cells produced up to 10-fold higher HBeAg titers and 20-fold higher HBsAg titers for #20 and #50 than HepG2/NTCP cells (Figure 3A–D). Therefore HepaRG cells most likely have a much higher susceptibility to sHBV infection than HepG2/NTCP cells even using crude serum samples and without FBS. Surprisingly, inoculating serum #27 without PEG produced only 0.4% of HBeAg in HepaRG cells than its presence, although for sample #40 the value was 54% (Figure 3A). In HepG2/NTCP cells the value ranged from 4% (#27) to 65% (#50) (Figure 3B). It is also interesting to note that the HBsAg/HBeAg ratio for HepaRG cells was reduced in Figure 3E than in Figure 1F (Table 3), possibly due to prolonged sample storage rather than PEG addition during inoculation. Another infection experiment performed 1 year earlier in HepG2/NTCP cells revealed much less PEG dependence for all the serum samples (Figure 3I). These findings implicate a labile host (human)-specific factor as a mediator of PEG-independent sHBV infection. Identification of such an elusive host factor would greatly advance our understanding of HBV entry into hepatocytes.
AUTHOR CONTRIBUTIONS
Shuping Tong, Jiming Zhang, and Jisu Li designed the study. Jing Li, Shiqi Chen, and Weicheng Xu performed the experiments. Jisu Li, Shuping Tong, and Jing Li analyzed the data. Jisu Li and Shuping Tong wrote the manuscript. All authors approved final version of the manuscript.
ACKNOWLEDGMENTS
The authors would like to thank all patients who participated in this study. This work was supported by NIH grants R01AI116639, R21AI142456, R21AI163819, R21AI166875, the National Natural Science Foundation of China (81672064, 81871640, 82172255, 82100634), the Fundamental Research Funds for the Central Universities (xzy012021057), the Nature Science Foundation of Shaanxi (2020JM-401) and the Major Science and Technology Special Project of China (2017ZX10202202 and 2017ZX10202203). Jing Li was supported by China Scholarship Council.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ETHICS STATEMENT
The study was conducted in accordance with the guidelines of the 1975 Declaration of Helsinki and was approved by the Institutional Ethics Committee for Human Studies at Huashan Hospital, Fudan University, Shanghai, China. Written informed consent was obtained from all patients.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.