Detection of H275Y oseltamivir resistance gene mutation among Influenza A(H1N1)pdm09 patients by allelic discrimination real-time RT-PCR
Smriti Krishna and Anup Jayaram are first authors.
Abstract
Influenza viruses can mutate genetically and cause a range of respiratory ailments. The H275Y mutation in the neuraminidase (NA) gene reduces the effectiveness of oseltamivir, a widely used drug for the treatment of Influenza A and B virus infection. The World Health Organization (WHO) recommends single-nucleotide polymorphism assays to detect this mutation. The present study aims to estimate the prevalence of H275Y mutation conferring oseltamivir resistance in Influenza A(H1N1)pdm09 virus among hospitalized patients from June 2014 to December 2021. Following the WHO protocol, allelic discrimination real-time RT-PCR was performed for 752 samples. Out of the 752 samples, 1 tested positive for Y275 gene mutation by allelic discrimination real-time RT-PCR. In samples of years 2020 and 2021, neither the H275 nor Y275 genotype was detected. Sequencing of the NA gene of all negative samples showed a mismatch between the NA sequence and the probes used in the allelic discrimination assay. Also, Y275 mutation was detected in only 1 sample from 2020. The prevalence of oseltamivir resistance was estimated as 0.27% among the Influenza A(H1N1)pdm09 patients during 2014−2021. The study highlights that the WHO-recommended probes for detecting H275Y mutation may not be useful to detect 2020 and 2021 circulating strains of Influenza A(H1N1)pdm09, emphasizing the need for continuous monitoring of mutations in the influenza virus.
1 INTRODUCTION
The influenza virus causes mild to severe respiratory infections resulting in 290 000−650 000 deaths globally.1 The Influenza viruses are spherical, filamentous, or pleomorphic with a diameter of ~100 nm. They have a lipid envelope with characteristic spikes called haemagglutinin and neuraminidase (NA).2 The fundamental activity of the NA protein involves cleaving of the sialic acids on the cell surface that cause the release of virions and infection of new cells.3 Oseltamivir carboxylate mimics the natural substrate of NA (sialic acid) and interacts with the active site, making Glu276 to rotate and bond with Arg224 which is stabilized by His275. These conformational alterations prevent the binding of sialic acid, and the subsequent release of the virus.4 The active sites of NA are known to be conserved, although mutations can affect these sites which may change virus transmissibility as well as drug sensitivity.5, 6 A point mutation in NA gene, H275Y, which is due to a nucleotide change of Cysteine (C) to Thymine (T) at 823 position resulting in an amino acid change of Histidine (H) to Tyrosine (Y) at 275 position conferring oseltamivir resistance is one of the most significant mutations.5, 7, 8
Antiviral usage has increased in recent times due to its effectiveness in reducing infections resulting in mild to moderate levels of resistance. Global Influenza Surveillance and Response System (GISRS) has deployed surveillance for influenza antiviral susceptibility testing. The GISRS states that phenotypic and genotypic assays are available to determine if an influenza virus is susceptible to NAIs. Genotypic assays comprise single-nucleotide polymorphism (SNP) detection, gene sequencing, and pyrosequencing.9 According to World Health Organization (WHO), the SNP based assay is currently used only for detection of H275Y mutation in influenza A(H1N1)pdm09 viruses and has been used in similar studies.10-14
Resistance to oseltamivir for A/(H1N1)pdm09 started to appear and was reported to affect immunocompromised individuals15 as well as patients who were treated with the drug.5 Resistance to oseltamivir has been detected as 2.14% in China,16 1.3% in Japan,17 1.2% in United States,18 1% in United Kingdom,15 and 0.74% in Indonesia.10 According to a study conducted by Trivedi et al.12 in India, the prevalence rate was 0.15% while Saha et al.19 and Nandhini et al.20 found that all the samples were sensitive to oseltamivir. In India, studies on the prevalence of oseltamivir resistant Influenza A(H1N1)pdm09 viruses are limited especially after the year 2015. Further, number of samples used in the previous studies are less and hence the possibility of detection of resistant virus is low. Thus, this study aims to detect the presence of H275Y mutation among Influenza A(H1N1)pdm09 patients from 2014 to 2021.
2 MATERIALS AND METHODS
2.1 Study population
Influenza A(H1N1)pdm09 positive samples archived at the Manipal Institute of Virology (MIV), from June 2014 to December 2021 were included in the study. Prevalence of oseltamivir resistance varies from 0% to 2% in India.12, 13 The sample size was estimated as 752, considering 2% prevalence (p = 0.02), 1% allowable margin of error (P/2 = 0.01 for rare disease), and 95% confidence interval using where, n is the number of samples, z is the statistic corresponding to the level of confidence (1.96).21
Out of 3023 influenza A(H1N1)pdm09 positive samples archived at MIV from 2014 to 2021, a total of 752 samples with Ct values <30 were selected from each year from different states of India by random sampling.
2.2 Allelic discrimination real-time RT-PCR
Influenza A(H1N1)−positive samples were tested used pooled sample testing strategy. Five positive samples with Ct value <30 were added to each pool making a total of 151 pools. Based on the sample volume required for manual extraction (140 μL), 30 μL of each sample was added to the pool and final elution volume was 60 μL. Nucleic acid was extracted from these pools using FavorPrep™ Viral DNA/RNA kit (Favorgen Biotech Corporation). The extracted nucleic acids were subjected to allelic discrimination real-time RT-PCR as per the WHO protocol7 using QuantiTect Virus Kit (Qiagen). NA-specific primers and probes for Influenza A(H1N1)pdm09 as recommended by WHO7 were purchased from Applied Biosystems, and reconstituted. Positive controls used for wild type—H275 (Cat No. FRS1168) and mutant type—H275Y (Cat No. FRS1169) were standard strains procured from International Reagent Resource.
Assay components and reaction conditions were standardized for allelic discrimination real-time RT-PCR. Briefly, reaction mix contained 5 µL of 5x QuantiTect Virus Master Mix, 0.5 µl of both forward and reverse primers (2 µM), 0.5 µL of H275 and Y275 probe each of 2 µM, 0.2 µL of QuantiTect Virus RT Mix, 12.8 µL of RNase free water, and 5 µL of template, making a total of 25 µL. Cycling conditions were programmed as a pre-read stage of 60°C for 1 min, reverse transcription process at 50°C for 20 min, followed by initial denaturation at 95°C for 10 min, with subsequent 45 cycles of amplification (denaturation at 95°C for 15 s and annealing and extension at 58°C for 45 s), and lastly, a post-read stage of 60°C for 1 min. PCR was performed using QuantStudio™ 6 Flex Real-Time PCR System (Applied Biosystems).
2.3 Modified Sanger sequencing
Sample homozygous for Y275 or negative for both H275 and Y275 targets in the allelic discrimination real-time RT-PCR were selected for sequencing. The partial fragment of NA gene was amplified using WHO-recommended primers. The desired product of 620 bp was extracted using GenElute™ Gel Extraction Kit (Sigma-Aldrich). Sequencing was done using BigDye Terminator (version 3.1) cycle sequencing kit (Applied Biosystems) and analyzed using 3500xL Genetic analyzer (Applied Biosystems) following the manufacturer's instructions.
2.4 Sequence analysis
Sequence analysis was done using MEGA X version 11 software. The reference sequence used for the analysis of 2014−2017 samples, Influenza A/Michigan/10/2009(H1N1)-like virus, was downloaded from National Center for Biotechnology Information. Reference sequences for 2020 and 2021 samples including WHO recommended vaccine strains sequences for Influenza A/Hawaii/70/2019(H1N1)-like virus and Influenza A/Wisconsin/588/2019(H1N1)-like viruses were downloaded from Global Initiative on Sharing All Influenza Data (GISAID).
3 RESULTS
3.1 Study population
Among the 752 samples selected from the different states of India for detection of H275Y gene mutation, the highest number of samples tested were from the state of Karnataka (40.4%, n = 304) followed by Tamil Nadu (19.6%, n = 148). The 2017 influenza season had the highest number of influenza cases hence a greater number of samples were selected for testing. The year-wise and geographic details of samples used in the study are summarized in Table 1.
| Sl No | Year | State | Number of samples | Number of pools | Allelic discrimination real time RT-PCR results | |||
|---|---|---|---|---|---|---|---|---|
| Homozygous H275 | Homozygous Y275 | Heterozygous H275/Y275 | Undetermined | |||||
| 1 | 2014 | Karnataka | 15 | 3 | 3 | 0 | 0 | 0 |
| 2 | 2014 & 2015 | Karnataka | 5 | 1 | 1 | 0 | 0 | 0 |
| 3 | 2015 | Karnataka | 23 | 6 | 6 | 0 | 0 | 0 |
| Kerala | 7 | |||||||
| 4 | 2015 & 2016 | Kerala | 1 | 1 | 1 | 0 | 0 | 0 |
| Assam | 3 | |||||||
| Gujarat | 1 | |||||||
| 5 | 2016 | Karnataka | 15 | 4 | 4 | 0 | 0 | 0 |
| Kerala | 1 | |||||||
| Tamil Nadu | 3 | |||||||
| Tripura | 1 | |||||||
| 6 | 2017 | Assam | 22 | 51 | 50 | 0 | 1 | 0 |
| Goa | 10 | |||||||
| Gujarat | 33 | |||||||
| Jharkhand | 19 | |||||||
| Karnataka | 46 | |||||||
| Kerala | 26 | |||||||
| Maharashtra | 27 | |||||||
| Odisha | 25 | |||||||
| Tamil Nadu | 47 | |||||||
| 7 | 2017 & 2018 | Tamil Nadu | 4 | 1 | 1 | 0 | 0 | 0 |
| Assam | 1 | |||||||
| 8 | 2018 | Assam | 2 | 35 | 35 | 0 | 0 | 0 |
| Gujarat | 6 | |||||||
| Karnataka | 56 | |||||||
| Kerala | 23 | |||||||
| Maharashtra | 9 | |||||||
| Tamil Nadu | 79 | |||||||
| 9 | 2018 & 2019 | Tamil Nadu | 2 | 1 | 1 | 0 | 0 | 0 |
| Odisha | 2 | |||||||
| Assam | 1 | |||||||
| 10 | 2019 | Assam | 3 | 38 | 38 | 0 | 0 | 0 |
| Goa | 4 | |||||||
| Gujarat | 8 | |||||||
| Jharkhand | 26 | |||||||
| Karnataka | 96 | |||||||
| Kerala | 18 | |||||||
| Maharashtra | 15 | |||||||
| Odisha | 4 | |||||||
| Tamil Nadu | 13 | |||||||
| 11 | 2020 | Karnataka | 35 | 7 | 0 | 0 | 0 | 7 |
| 12 | 2020 & 2021 | Goa | 1 | 1 | 0 | 0 | 0 | 1 |
| Karnataka | 3 | |||||||
| Kerala | 1 | |||||||
| 13 | 2021 | Karnataka | 10 | 2 | 0 | 0 | 0 | 2 |
| Total | 752 | 151 | 140 | 0 | 1 | 10 | ||
3.2 Allelic discrimination real-time RT-PCR
Endpoint fluorescent signals were analyzed and the threshold was adjusted manually. The results were categorized as homozygous wild-type (H275), homozygous mutant-type (Y275), and heterozygous type (H275/Y275).
Out of the 151 pools that were tested for H275Y mutation, 140 pools were homozygous H275 and only 1 pool showed amplification for both H275 and Y275 (heterozygous type) and 10 pools were negative for both H275 and Y275 (Figure 1 and Table 1).
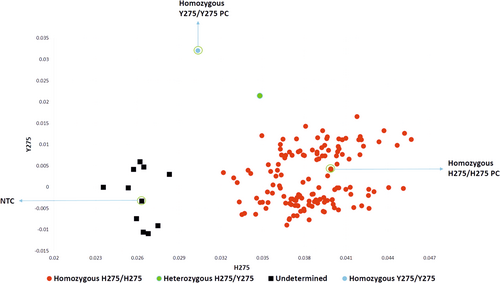
The five samples of the heterozygous positive pool were tested individually and one sample was detected as homozygous mutant-type (Y275). The prevalence rate of oseltamivir resistance from 2014 to 2019 was found to be 0.13%.
3.3 Modified Sanger sequencing
Sequence analysis of homozygous Y275 sample in comparison to reference strains showed a nucleotide change of CAC to TAC at 823 position resulting in an amino acid change from H to Y at 275 position, confirming the presence of H275Y mutation.
Fifty samples from the 10 undetermined pools of 2020 and 2021 were sequenced and sequence analysis showed no nucleotide change at 823 position for 49 samples, however one sample from 2020 showed nucleotide change of CAC to TAC at 823 position resulting in an amino acid change from H to Y at 275 position. The prevalence rate of oseltamivir resistance from 2014 to 2021 was found to be 0.27%. All the nucleotide sequences (510 bp) from this study were deposited in GenBank with accession numbers from OP737518 to OP737538 and OQ363464 to OQ363489. Among these, OP737518 and OQ363483 had the codon change of CAC to TAC with amino acid mutation of H to Y.
Sequence alignment of all 2020 and 2021 samples with the reference sequences and WHO recommended probe showed a mutation of GAG to GAA at 831 position. This nucleotide mismatch with probe could have possibly resulted in negative results for both H275 and Y275 targets in allelic discrimination PCR.
Further, nucleotide BLAST analysis of the probe binding region using GISAID analysis tool showed that the nucleotide change of G to A at 831 position has been reported since end of 2019 (GISAID No. EPI_ISL_16071680) in different regions of the world and this nucleotide change is also observed in the WHO recommended vaccine strain for 2021−2022—Northern hemisphere (Figure 2).

4 DISCUSSION
Oseltamivir is the most widely used antiviral for the treatment of influenza infections in India, therefore it is important to evaluate its susceptibility and effectiveness. H275Y mutation leading to oseltamivir resistance has a prevalence of 1%−2% worldwide and less than 1% in India. Gohil et al.22 conducted a study for the surveillance of the influenza virus in Mumbai and found that all the samples were sensitive to oseltamivir. Similar studies by Sahu et al.13 during 2009−2015 and Trivedi et al.12 during 2015−2016 demonstrate the prevalence of oseltamivir resistance as 1.4% and 0.15%, respectively.
In India, the number of published studies on oseltamivir resistance after the year 2015 are sparse. Hence, this study was conducted to estimate the prevalence of oseltamivir resistance among Influenza A(H1N1)pdm09 patients from 2014 to 2021 using allelic discrimination real-time RT-PCR to identify H275Y gene mutation.
The current study, estimates prevalence rate of oseltamivir resistance as 0.27% among hospitalized patients that was found to be similar to previous studies from India.12-14, 19, 20 Based on the results, it can be concluded that oseltamivir resistance is still less than 1% in India. Out of two resistant samples, one was from the year 2017, collected from 26-year-old male, with 2 days history of fever, cough, coryza, red eye, retro-orbital pain, myalgia, and mild gastrointestinal symptoms. He had received oseltamivir treatment and there was no history of previous influenza infection or oseltamivir treatment. However, the follow-up report showed the patient had recovered and was healthy. The second resistant sample (collected in 2020) was from a 55-year old male, with 2 days history of fever, cough, difficulty in swallowing, breathlessness, and gastrointestinal symptoms. The second patient had also received oseltamivir treatment with no previous history of infection or resistance. However, the patient's condition deteriorated and hypoxia worsened, secondary bacterial infection was also detected. Due to the patient's financial restraints he was discharged against medical advice. A few studies from Hong Kong,23 Vietnam,24 and United States,18 have reported oseltamivir resistance in patients without prior history of treatment. A similar finding was reported by Tandel et al.25 in a study conducted in Madhya Pradesh, India, where drug resistance was detected in patients without any previous exposure. Oseltamivir resistant and oseltamivir-susceptible viral infections are reported to be clinically similar, with the exception of those who are immunocompromised or have comorbid conditions.26
Allelic discrimination real-time RT-PCR is a rapid SNP detection method that detects H275Y mutation and has been recommended by WHO. In the current study, pooled sample testing strategy was used to detect H275Y gene mutation. Pooled sample testing strategy has been used to detect viruses such as hepatitis B virus, human immunodeficiency virus, influenza virus among others.27 This testing strategy with pool sizes of 5−10 samples has also been widely used to increase testing capacity and conserve resources during upsurge testing periods such as the on-going COVID-19 pandemic.28 A study by Shukla et al. showed that the average change in Ct for pool of 5 was +2.56 and +3.38 for pool of 10.28 To avoid false positives and increase efficiency of allelic discrimination real-time RT-PCR, pools of 5 influenza A(H1N1)pdm09 positive samples with Ct <30 were tested for SNP detection.
In this study, the samples of 2020 and 2021 were negative for both wild-type and mutant-type. Based on sequencing results, it was confirmed that 49 samples did not have the H275Y mutation, but one sample was detected with H275Y mutation. On further analysis of sequences, an additional nucleotide mutation was detected at the probe binding site that might have contributed to negative results for both H275 and Y275. Allelic discrimination assays are very specific and a single mismatch between the template and the probe sequence will affect its hybridization.29
Further, nucleotide BLAST analysis of probe binding region showed that these mutations might have emerged toward the end of 2019 (GISAID No. EPI_ISL_16071680, unpublished data) and has been detected in many countries since its emergence. The WHO recommended vaccine strains sequence for Northern Hemisphere—2021−2022 also documents this change (GISAID No. EPI_ISL_404527). Although, the nucleotide change at 831 position results in a synonymous mutation and does not affect drug binding, the mismatch at the probe binding site is a major concern since these probes are recommended by WHO for detection of the substitution at amino acid 275 in the NA of influenza A(H1N1)pdm09 viruses. The WHO, allelic discrimination real-time RT-PCR is considered as a rapid and convenient method and is widely used for screening of mutations associated with drug resistance.12 The detection of nucleotide change in probe binding site of 2020 and 2021 samples strengthens the fact that considering the high mutation rates among influenza A viruses the WHO protocol recommendations needs to be revised periodically to avoid false results.
5 CONCLUSION
The evolving nature of influenza viruses poses a challenge for developing vaccines and antivirals. The NA inhibitors against influenza viruses are effective and used extensively. Any mutation in the drug's binding site could result in a decrease in the drug's effectiveness and eventual discontinuation. This study illustrates a 7-year profile of H275Y gene mutation leading to oseltamivir resistance among Influenza A(H1N1)pdm09 patients from 2014 to 2021. The result shows a lower prevalence of 0.27% which indicates that oseltamivir carboxylate can be continued to be used as a therapeutic. The negative results obtained in the samples of 2020 and 2021 using allelic discrimination real-time RT-PCR following WHO protocol highlights the necessity of monitoring the mutations in the influenza virus and implementing it in the respective assays so as to avoid any false negative results.
AUTHOR CONTRIBUTIONS
Anitha Jagadesh conceived, and guided the research work. Smriti Krishna performed most of the experiments. Anup Jayaram contributed to data processing and analysis. Ujwal Shetty contributed to performing sequencing. Prasad Varamballi contributed in data analysis. Chiranjay Mukhopadhyay provided insightful discussion. All authors have read and approved the manuscript
ACKNOWLEDGMENTS
We thank all the staff and students of Manipal Institute of Virology, Manipal Academy of Higher Education for their constant support. Funding was by Manipal Institute of Virology, Manipal Academy of Higher Education, Manipal, Karnataka, India.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ETHICS STATEMENT
The study was reviewed and approved by the Institutional Ethical Committee (IEC), Kasturba Medical College, and Kasturba Hospital (Approval ID-IEC2: 179/2022).
Open Research
DATA AVAILABILITY STATEMENT
Data supporting the current study is available from the corresponding author upon request.




