Infectivity of pseudotyped SARS-CoV-2 variants of concern in different human cell types and inhibitory effects of recombinant spike protein and entry-related cellular factors
Abstract
Since the report of the first COVID-19 case in 2019, SARS-CoV-2 variants of concern (VOCs) have continued to emerge, manifesting diverse infectivity, evasion of host immunity and pathology. While ACE2 is the predominant receptor of SARS-CoV-2, TMPRSS2, Kim-1, NRP-1, CD147, furin, CD209L, and CD26 have also been implicated as viral entry-related cofactors. To understand the variations in infectivity and pathogenesis of VOCs, we conducted infection analysis in human cells from different organ systems using pseudoviruses of VOCs including Alpha, Beta, Gamma, and Delta. Recombinant spike S1, RBD, ACE2, Kim-1, and NRP-1 proteins were tested for their ability to block infection to dissect their roles in SARS-CoV-2 entry into cells. Compared with wild type SARS-CoV-2 (WT), numerous VOCs had significant increases of infectivity across a wide spectrum of cell types. Recombinant ACE2 protein more effectively inhibited the infection of VOCs including Delta and Omicron (BA.1 and BA.2) than that of WT. Interestingly, recombinant S1, RBD, Kim-1, and NRP-1 proteins inhibited the infection of all pseudoviruses in a manner dependent on the levels of ACE2 expression in different cell types. These results provide insights into the diverse infectivity of SARS-CoV-2 VOCs, which might be helpful for managing the emergence of new VOCs.
1 INTRODUCTION
The COVID-19 pandemic started in November 2019 and has since persisted globally.1 Despite the development of effective vaccines against the original wild type (WT) strain of the causal agent SARS-CoV-2, the virus has continued to evolve, generating numerous variants of concern (VOCs) including Alpha (B.1.1.7), Beta (B.1.351), Gamma (P1), Delta (B.1.617.2), and Omicron (B.1.1.529).2, 3 The Omicron variant was first reported in November 2021 in Botswana and South Africa,4 and five major lineages including BA.1, BA.2, BA.3, BA.4, and BA.5 have been documented so far and new ones have continued to emerge.5-7 Compared with the WT, most of the VOCs contain several mutations.6 One mutation is D614G, which is responsible for the enhanced infectivity and more effective evasion of host immunity observed in VOCs.6, 8
Clinically, SARS-CoV-2 causes symptoms or pathology in multiple organs from different human systems.9, 10 Meanwhile, SARS-CoV-2 RNAs and proteins have been detected in multiple types of cells and organs from COVID-19 patients.11-13 Experimentally, SARS-CoV-2 WT pseudovirus can infect multiple types of cells from different human systems including respiratory, urinary, immune, digestive, and reproductive.14 However, patients infected by different VOCs manifest diverse symptoms and pathology, possibly suggesting tropisms for different organs and cell types.15
Angiotensin converting enzyme 2 (ACE2) is the predominant receptor of SARS-CoV-2.16 The expression level of ACE2 protein varies in different tissues or cells.12, 14 Tissues from respiratory, urinary and digestive systems that are more susceptible to SARS-CoV-2 infection have relatively higher ACE2 expression levels.14 However, some tissues or cells with low ACE2 expression levels such as olfactory cells are also susceptible to SARS-CoV-2 infection.17 Subsequent works have identified other cellular proteins involved in SARS-CoV-2 infection, possibly by serving as alternative receptors, co-receptors or entry regulatory cofactors. These include type II membrane serine proteases (TMPRSS2), kidney injury molecule-1 (Kim-1), neuropilin-1 (NRP-1), cluster of differentiation 147 (CD147), dipeptidyl peptidase-4 (DPP4 or CD26), C-type lectins (DC-SIGN or CD209, l-SIGN or CD209L, LSECtin or CLES4G, ASGR1, and CLEC10A), and furin, and so forth.18
SARS-CoV-2 spike (S1) protein and entry-related proteins are potential targets for vaccines or therapeutic antibodies.19, 20 Soluble recombinant ACE2 can effectively inhibit SARS-CoV-2 infection by blocking the interaction of receptor-binding domain (RBD) of the spike protein with cellular ACE2.14, 21 Both recombinant S1 and RBD proteins can block SARS-CoV-2 infection but RBD exhibits higher binding affinity to the ACE2 receptor and can more effectively inhibit SARS-CoV-2 infection.22 Similar to SARS-CoV-2 nucleocapsid protein, S1 protein has been extensively used in the development of diagnostic methods.23-25
To understand the tropisms and pathology associated with VOCs, we examined the infectivity of pseudoviruses of SARS-CoV-2 WT and VOCs in different types of cells from various human organ systems. The inhibitory effects of recombinant S1 and RBD proteins, and recombinant cellular proteins ACE2, Kim-1 and NRP-1 on the pseudoviruses were further examined in cells with high (Huh7) and low (A549) ACE2 protein levels.
2 MATERIALS AND METHODS
2.1 Cell lines and cell culture
JHU-029, NCI-H460, NCI-H322, NCI-H520, A549, HLBEC, HSAEC, 768-O, A498, 769-P, Caki-1, ACHN, HRC45, HRC63, HRC59, BCP-1, BC-3, BJAB, THP-1, Huh-7, PCI-13, UD-SCC-2, HUVEC, T47D, and MCF-7 cells were cultured as previously described.14, 19 We tested the infectivity of pseudoviruses of SARS-CoV-2 WT and VOCs in these 25 cell lines. HEK293T cells were used for packaging of pseudoviruses, while HEK293T/hACE2 cells were used to test the inhibitory effects of different recombinant proteins on infection of pseudoviruses. All the cells were maintained at 37°C with 5% CO2.
2.2 Plasmids for packaging of pseudoviruses
Plasmids expressing the spike proteins of SARS-CoV-2 WT and VOCs (Alpha, Beta, Gamma, Delta, and Omicron) were used for packaging of the respective pseudoviruses as previously described.19, 26, 27 Plasmids expressing the spike protein of SARS-CoV-2 VOCs were gifts from David Nemazee,27, 28 except the Omicron S protein gene plasmid, which was a gift from Dr. Tongqing Zhou.29 Briefly, the spike genes were cloned from the original plasmids into pcDNA3.1(+) by restriction digestion and T4 ligation. Plasmid pNL4-3. Luci.R-E- was used as pseudovirus backbone in pseudovirus packaging. Plasmid pcDNA3.1(+) was used in the packaging as a blank negative control while plasmid pMD2.G expressing the VSV-G envelope protein was used as a positive control.
2.3 Western-blotting
The protein expression levels of ACE2, TMPRSS2, Kim-1, and NRP-1 in different cell lines were previously examined by Western-blotting, quantified with the ImageJ software, and used in the heatmap analysis using the Graphpad 9 software.14
2.4 Packaging of pseudoviruses and infection assay
Pseudoviruses of SARS-CoV-2 WT and VOCs were packaged as previously described.14, 19 Briefly, plasmid pNL4-3. Luci.R-E- and each of the spike protein plasmids were cotransfected into HEK293T using jetOPTIMUS transfection reagent (Polyplus) at a 2:1 ratio. At 2 day posttransfection, the supernatant was collected and centrifuged at 4000 rpm for 5 min. The supernatant was passed through a 0.45 μm filter, aliquoted and stored in a −80°C freezer. Pseudoviruses were titrated before infection as previously described.14
2.5 Blocking assay
The inhibitory effects of recombinant proteins S1, RBD, ACE2, Kim-1, and NRP-1 (Sino Biological) on the infectivity of pseudoviruses in Huh7 and A549 cells were examined.14 Briefly, the tested pseudovirus was incubated with a recombinant protein at 2 µg/ml for 30 min and added to Huh7 or A549 cells. The media was changed after overnight incubation. At 60 h postinfection (hpi), cells were lysed and assayed for luciferase activity. Infection was conducted in triplicate and the experiments were repeated three times.
2.6 Determine the IC50 of recombinant ACE2 protein's inhibitory effects on pseudoviruses
The IC50 of ACE2 protein's inhibitory effects on pseudoviruses of SARS-CoV-2 WT, and Alpha, Beta, Gamma, Delta, and Omicron (BA.1 and BA.2) VOCs were examined. The inhibitory assay was conducted as previously described with minor modifications.14 SARS-CoV-2 pseudoviruses were incubated with two-fold serial diluted recombinant ACE2 protein in PBS starting at 4 μg/ml. The media was changed after overnight incubation. Cells were lysed and the luciferase activity was measured at 60 hpi.14 Infection was conducted in triplicate and the experiments were repeated three times.
2.7 Statistical analysis
Results were analyzed with GraphPad Prism 9. One-way analysis of variance (ANOVA) was performed if multiple samples were involved. p < 0.05 was considered statistically significant.
3 RESULTS
3.1 Enhanced infectivity of pseudoviruses of SARS-CoV-2 VOCs in cells from different human organ systems
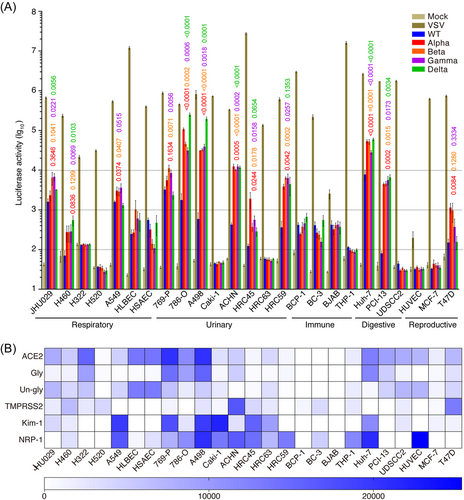
We used pseudoviruses of SARS-CoV-2 WT and VOCs including Alpha, Beta, Gamma, and Delta to infect different cell types from 5 organ systems including respiratory, urinary, immune, digestive, and reproductive systems. We detected minimal activities with the pseudovirus packaged with the vector control pcDNA3.1(+) alone (Mock) but broad strong activities with the positive control pseudovirus packaged with the VSV-G envelope protein in all cell lines tested (Figure 1A). As previously reported,14 WT pseudovirus infected different cell types from all 5 organ systems including JHU029, H460, H322, A549, HLBEC, and HSAEC from the respiratory system; 769-P, 786-O, A498, ACHN, HRC45, and HRC59 from the urinary system; BCP-1, BC-3, BJAB, and THP-1 from the immune system; Huh-7 from the digestive system; and T47D from the reproductive system. Compared to WT pseudovirus, Alpha, Beta and Gamma pseudoviruses exhibited significantly higher infectivity in JHU029, H460, A549, 769-P, 786-O, A498, ACHN, HRC45, HRC59, Huh-7, PCI-13, and T47D cells. Delta pseudovirus also had higher infectivity than the WT pseudovirus had in these cells except for A549, 769-P, and T47D cells, in which WT and Delta pseudoviruses had similar infectivity. Compared to WT pseudovirus, strong increases of infectivity of pseudoviruses of all VOCs in the range of 3- to 300-fold were observed in 786-O, A498, ACHN, HRC59, and Huh-7 cells. Interestingly, H460 and PCI-13 were refractory to WT pseudovirus infection but were permissive to pseudoviruses of all VOCs tested, showing robust activities. Together, these results suggest that pseudoviruses of all VOCs are generally more infectious than the WT pseudovirus. Furthermore, cell types that were highly permissive to all pseudoviruses (WT and VOCs) including JHU029, A549, 769-P, 786-O, A498, ACHN, HRC59, Huh-7, and PCI-13 cells also had strong expression levels of at least one or several entry-related factors including ACE2, TMPRSS2, Kim-1, and NRP-1 proteins (Figure 1B). Interestingly, among these cell lines, relatively lower levels of ACE2 were detected in A549, ACHN, and HRC59 than other permissive cells suggesting the possible usage of alternative receptor(s) or involvement of entry-related cofactor(s) other than ACE2 protein in these cells (Figure 1B).
3.2 Inhibition of infection of pseudoviruses of SARS-CoV-2 WT and VOCs by recombinant entry-related proteins
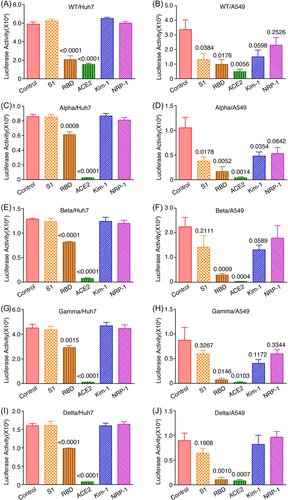
Recombinant proteins of SARS-CoV-2 entry-related proteins can block SARS-CoV-2 infection.14, 30 We examined the inhibitory effects of recombinant S1, RBD, ACE2, Kim-1, and NRP-1 proteins on the infection of pseudoviruses of VOCs in Huh-7 and A549 cells. Huh-7 cells have a high while A549 cells have a low expression level of ACE2 protein.14 Both cell lines have high expression levels of Kim-1 and NRP-1 proteins.14 Infectivity of pseudoviruses of WT and VOCs was inhibited by recombinant RBD but not S1 protein in Huh7 cells (Figure 2A,C,E,G,I). Recombinant RBD protein also manifested stronger inhibitory effects on all pseudoviruses in A549 cells (Figure 2B,D,F,H,J). Nevertheless, recombinant S1 protein had some inhibitory effects on all pseudoviruses in A549 cells. These results could be due to the dose-dependent competitive nature of the blocking effects on viral binding to the ACE2 receptor of recombinant S1 and RBD proteins. As expected, recombinant ACE2 protein manifested strong inhibitory effects on the infectivity of all pseudoviruses in both Huh-7 and A549 cells (Figure 2). While recombinant Kim-1 and NRP-1 proteins had no detectable inhibitory effect on the infectivity of all pseudoviruses in Huh-7 cells, they manifested some low levels of inhibitory effects on these pseudoviruses in A549 cells (Figure 2). These results support the important role of ACE2 protein in SARS-CoV-2 infection reported in other studies.31, 32 Both Kim-1 and NRP-1 proteins might contribute to SARS-CoV-2 infection, particularly in cells that have a low expression level of ACE2 protein.
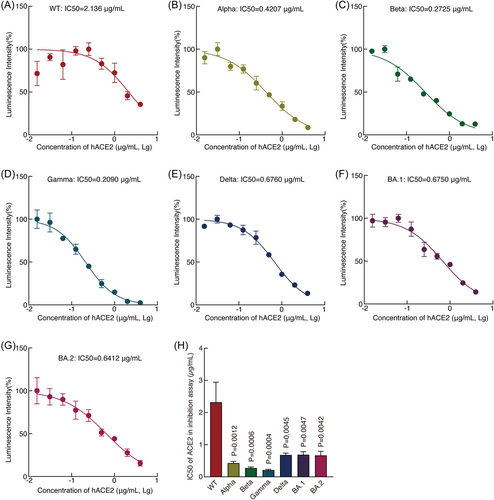
Since recombinant ACE2 protein had the strongest inhibitory effects on all pseudoviruses, we determined its IC50s on pseudoviruses. Recombinant ACE2 protein had an IC50 of 2.1360 μg/ml against the WT pseudovirus (Figure 3A). Much lower IC50s were observed for pseudoviruses of all VOCs including Omicron BA.1 and BA.2 pseudoviruses ranging from 0.2090 for the Gamma pseudovirus to 0.6760 for the Delta pseudovirus (Figure 3), suggesting recombinant ACE2 protein's much higher inhibitory effects against pseudoviruses of VOCs than that of WT.
4 DISCUSSION
SARS-CoV-2 has a broad tropism for diverse cell types and organs.11, 12, 14 Multiple mutations have been identified in the spike gene of VOCs, which enhance the binding affinity between the spike protein and ACE2 protein, the predominant binding receptor of SARS-CoV-2.33 In particular, mutation D614G is present in almost all VOCs, and is responsible for the enhanced infectivity and evasion of host immunity.6, 8 Several waves of VOCs have been reported so far, most of which have manifested higher transmission rates than the original (WT) virus, leading to the rapid rise of COVID-19 cases all over the world.34 We hypothesized that the faster spread of VOCs in the human population might be due to their broader permissiveness and higher infectivity in cells and organs. We performed comparative analyses of pseudoviruses of WT and VOCs in 25 cell lines consisting of different cell types from 5 human organ systems (Figure 1A). Compared with the SARS-CoV-2 WT, pseudoviruses of VOCs infected more cell lines and had enhanced infectivity, suggesting that they indeed have broader tropism and permissiveness. These results could explain the fast spread of some VOCs in the human population. In agreement with these results, Alpha, Beta, and Gamma VOCs induced a wider range of symptoms, expanded systemic infection to the gastrointestinal tract, elicited depletion of natural killer cells, and triggered variant-specific cytokine production patterns compared to the early 2020 isolate EU-1 in human ACE2 transgenic mice.35 Remarkably, VOCs were shown to use the murine ACE2 as the entry receptor for infection in the lungs of wild-type mice,35 suggesting likely evolved binding promiscuity of the spike protein of VOCs to its receptor. Furthermore, the Omicron variants BA.1 and BA.2 had more robust replication than WT and the Delta variant in bronchial tissues at 37°C, and BA.2 replicated more efficiently in nasal and bronchial tissues at 33°C than WT, and Delta and Omicron BA.1 variants.36 In this study, we included Omicron BA.1 and BA.2 pseudoviruses in our analysis. However, over a dozen Omicron subvariants have since emerged.6 A more comprehensive study with these Omicron subvariants could further delineate their cell tropism and infectivity leading to better understanding of their transmission and pathogenesis.
We acknowledge the limitation of using pseudovirus for examining viral tropism and infectivity. While pseudovirus is a powerful tool for vaccine and therapeutic antibody evaluations, it neither recapitulates the entire viral life cycle and nor has multiple rounds of infection. Hence, it remains important to validate results obtained with pseudovirus with wild-type virus-based assay.37
While ACE2 is the predominant SARS-CoV-2 receptor, numerous other proteins including TMPRSS2, Kim-1, NRP-1, CD26, CD147, CD206, and furin are also involved in SARS-CoV-2 cell entry.38 The expression levels of these proteins in different cell types and organs vary.14 In fact, cells that are more permissive to pseudoviruses of VOCs also have relatively higher expression levels of one or several entry-related proteins (Figure 1). The enhanced infectivity of VOCs in some cell types and organs suggest that mutations in the spike gene might have optimized the interactions between the spike protein and one or more entry-related proteins. Indeed, compared to WT, the transmissibility of Alpha, Beta, Gamma, and Delta has increased.39-42 While patient tissue analysis or tropism studies indicates that SARS-CoV-2 WT has multiorgan tropism, the tropism of VOCs has rarely been reported.11, 12 Our results of enhanced infectivity of VOCs in different cell types from numerous organs compared with that of WT support these observations. In particular, we observed enhanced infectivity of VOCs in cells with a low level of ACE2 protein (Figure 1). It is unclear whether the cell entry mechanism of VOCs into cells with a low level of ACE2 protein remains the same. Nevertheless, cell-to-cell transmission of SARS-CoV-2 has been reported.43 This mechanism likely reduces the ACE2 dependency for viral entry, thus providing an alternative infection mechanism in cells with low level of ACE2, particularly for those that have close cell-cell contact or form tight junctions. Furthermore, other regulatory cofactors could also play crucial roles in SARS-CoV-2 infection in cells with a low level of ACE2 protein.44
Efficient SARS-CoV-2 infection depends on the interactions between the spike protein and the host receptor(s) as well as other entry-related cofactors. While the SARS-CoV-2 receptor ACE2 protein is broadly expressed in host tissues, other studies have also shown SARS-CoV-2 infection in tissues with a low expression level of ACE2 protein.44 There are likely alternative receptor(s) and entry-related cofactors that might mediate SARS-CoV-2 entry into cells, particularly in tissues with a low expression level of ACE2 protein.18 Study on the expression profile of SARS-CoV-2 entry-related cofactors could help understand the tropism and infectivity of SARS-CoV-2. We have previously analyzed the mRNA and protein expression levels of ACE2, TMPRSS2, Kim-1, and NRP-1 in 25 cell lines from respiratory, urinary, digestive, reproductive, and immune systems.14 The protein expression levels of ACE2 and NRP-1 were shown to be positively associated with the infectivity of SARS-CoV-2 WT.14
In the blocking assay, recombinant ACE2 protein effectively inhibited the infection of pseudoviruses of WT and all VOCs in cells with both high and low expression levels of ACE2. However, recombinant ACE2 protein had a higher IC50 for WT than those of VOCs (Figure 3). It is worth noting that soluble ACE2 at physiological concentration could facilitate the virus infection.45 However, higher soluble ACE2 protein at 25 ug/ml and 100 ug/ml totally inhibited SARS-CoV-2 infection,21 which is consistent with our results. The concentration of soluble ACE2 protein used in these studies are much higher than that at physiological status. Interestingly, recombinant S1 and RBD proteins can more efficiently inhibit the infection of pseudoviruses of WT and all VOCs in cells with a lower than a higher expression level of ACE2 protein (Figure 2). Furthermore, weak inhibitory effects were observed with recombinant Kim-1 and NRP-1 proteins against the pseudoviruses but only in cells with a lower expression level of ACE2. These results suggest that the ACE2 protein is the dominant receptor for both WT and VOCs. However, in cells that express a lower level of ACE2 protein, other entry-related factors might partially compensate for its function in SARS-CoV-2 infection, which could provide VOCs with broader tropism and higher transmissibility.41, 46 Consistent with the observed broader infectivity, VOCs also manifest wider pathological symptoms.35
In summary, the increased infectivity of SARS-CoV-2 VOCs in different cell types from diverse organ systems might contribute to their increased infectiousness and transmissibility. Recombinant ACE2 protein can more efficiently inhibit the infection of VOCs than WT, which could be due to the enhanced affinity between the ACE2 and spike proteins of VOCs. The fact that recombinant Kim-1 and NRP-1 proteins manifest inhibitory effects on the infection of pseudoviruses of WT and VOCs but only in cells with a lower expression level of ACE2 suggest their involvement in SARS-CoV-2 infection in a broader range of organs and tissues with diverse expression levels of ACE2 protein.
AUTHOR CONTRIBUTIONS
Conceptualization, planning, and management: Shou-Jiang Gao. Laboratory experimental design: Shou-Jiang Gao and George Fei Zhang. Laboratory execution of experiments, and data acquisition, analysis, and interpretation: George Fei Zhang, Wen Meng, Luping Chen, Ling Ding, Shenyu Sun, Xian Wang, Yufei Huang, Haitao Guo, and Shou-Jiang Gao. Drafting and revision of the manuscript: George Fei Zhang and Shou-Jiang Gao. Manuscript editing and approval: George Fei Zhang, Wen Meng, Luping Chen, Ling Ding, Shenyu Sun, Xian Wang, Yufei Huang, Haitao Guo, and Shou-Jiang Gao.
ACKNOWLEDGMENTS
This study was in part supported by UPMC Hillman Cancer Center Startup Fund and Pittsburgh Foundation Endowed Chair in Drug Development for Immunotherapy to S. J. G.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
Data that support the findings of this study are available from the corresponding author upon reasonable request.




