Haplotypic variants of COVID-19 related genes are associated with blood pressure and metabolites levels
Abstract
The genetic association of coronavirus disease 2019 (COVID-19) with its complications has not been fully understood. This study aimed to identify variants and haplotypes of candidate genes implicated in COVID-19 related traits by combining the literature review and pathway analysis. To explore such genes, the protein-protein interactions and relevant pathways of COVID-19-associated genes were assessed. A number of variants on candidate genes were identified from Genome-wide association studies (GWASs) which were associated with COVID-19 related traits (p ˂ 10-6). Haplotypic blocks were assessed using haplotypic structures among the 1000 Genomes Project (r2 ≥ 0.8, D′ ≥ 0.8). Further functional analyses were performed on the selected variants. The results demonstrated that a group of variants in ACE and AGT genes were significantly correlated with COVID-19 related traits. Three haplotypes were identified to be involved in the blood metabolites levels and the development of blood pressure. Functional analyses revealed that most GWAS index variants were expression quantitative trait loci and had transcription factor binding sites, exonic splicing enhancers or silencer activities. Furthermore, the proxy haplotype variants, rs4316, rs4353, rs4359, and three variants, namely rs2493133, rs2478543, and rs5051, were associated with blood metabolite and systolic blood pressure, respectively. These variants exerted more regulatory effects compared with other GWAS variants. The present study indicates that the genetic variants and candidate haplotypes of COVID-19 related genes are associated with blood pressure and blood metabolites. However, further observational studies are warranted to confirm these results.
1 INTRODUCTION
Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS‑CoV‑2), which is classified as RNA viruses. High infection rates and the emergence of numerous clinical manifestations are the hallmarks of COVID-19 infection. Many international and observational studies have focused on these issues over the past 2 years.1-4 These studies will help to better understand COVID-19 infection, reduce its complications, and find effective treatments. The respiratory tract, blood metabolites, and cardiovascular system are commonly affected by COVID-19 infection.
Many studies have shown the impact of different genetic variants on blood metabolites, as well as respiratory or cardiovascular diseases.5-7 The Genome-wide association study (GWAS) is one of the most important approaches used for identifying the association between genetic variants and complex disorders. The identification of disease-causing genes to understand the genetic function, communication, and pathways has always been a challenge in genetic studies. Several bioinformatics studies have been performed to identify and predict the effect of genetic variants in a number of diseases.8, 9
Several studies have been performed to characterize variants and haplotype involved in COVID-19 related traits.10, 11 The understanding of these factors will help to seek correlations between COVID-19 infection and its associated characteristics and genetic variants, as well as genetically related mechanisms, for effective targeted treatments. This study hypothesized that previously identified polymorphisms by GWAS could provide major clues about the genetic association of COVID-19 and its associated diseases or traits. Therefore, this study aimed to find genetic variants and haplotypes among candidate genes associated with COVID-19 related traits by combining the literature review and pathway analysis.
2 METHODS
This study was based on the combinatory assessment of the literature reviews and pathway analysis approaches. To achieve this goal, candidate genes associated with COVID-19 infection were identified by the literature review and pathway analysis, and then previous GWAS results and their significant variants associated with the candidate genes were identified from the National Human Genome Research Institute-European Bioinformatics Institute(NHGRI-EBI) GWAS catalog (https://www.ebi.ac.uk/gwas/). Finally, the haplotypic blocks associated with these candidate variants were characterized, and their functional relevance was analyzed. An overview of the workflow is shown in Figure 1. The detailed method is described in the following sections.
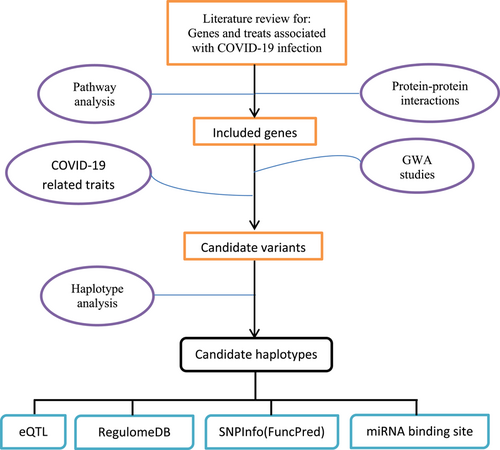
3 CANDIDATE GENES ASSOCIATED WITH COVID-19 INFECTION
To investigate the significant genes associated with COVID-19, a literature review was performed on experimental human studies available in PubMed. We used “COVID-19,” “gene,” “expression,” “polymorphism,” and their equivalent terms to identify the main genes whose expression levels or one of their polymorphic variants were correlated with coronavirus infection, mortality, or severity of disease in both male and female adult patients. To this aim, the kyoto encyclopedia of genes and genomes (KEGG) pathways (https://www.genome.jp/kegg/pathway.html)12 and protein−protein interactions (https://string-db.org/)13 were assessed according to the highest confidence score (0.9), coexpression, experimentally determined, curated databases, and text mining interactions. Based on the literature review and pathway analysis, a list of candidate genes associated with COVID-19 infection was obtained.
4 GWAS VARIANTS ASSOCIATED WITH COVID-19 RELATED TRAITS
The most important COVID-19 related traits listed in previous studies were identified. GWAS variants associated with these identified traits were obtained from GWAS results listed in the EBI-NHGRI GWAS catalog (https://www.ebi.ac.uk/gwas/). The catalog format was tab-separated values containing the results of 3875 GWA studies and 111 406 variants. Based on traits, SNPs, and p values, a data set of index variants (GWAS statistically significant variants(p ˂ 10−6)) in candidate genes associated with COVID-19 infection was obtained from the EBI-NHGRI GWAS catalog. The sample size of the studied populations was obtained from the sum of the initial and replication of the sample size. Then, GRCh38 genomic positions of these variants were extracted from single nucleotide polymorphism database (https://www.ncbi.nlm.nih.gov/snp/). The GRCh38 genomic position of candidate genes associated with COVID-19 infection (identified based on statements previously mentioned) was obtained from the National Center for Biotechnology Information gene (https://www.ncbi.nlm.nih.gov/gene). Then, variants whose genomic positions were placed inside the candidate genes were characterized using the Galaxy web-based platform.14
5 HAPLOTYPE OF CANDIDATE VARIANTS
Candidate variants associated with COVID-19 related traits were used to obtain haplotype blocks. These variants were integrated with the data obtained from population-specific haplotypic structures from the 1000 Genomes Project. Haplotypes associated with each COVID-19 related trait were separately analyzed using HaploReg v4.1. For this purpose, all GWAS variants associated with COVID-19 related traits (identified in the previous step) were included in the analysis. Variants with high correlation ratios (r2 ≥ 0.8, D′ ≥ 0.8) were recorded for COVID-19 and its related traits influencing haplotype blocks.
6 FUNCTIONAL ANALYSIS
To assess the impact of variants in haplotype blocks, their functional effects were investigated. A comprehensive set of micro RNAs (miRNA) targets were prepared to find variants located in miRNA binding sites, including the information about the binding sites confirmed by experimental studies, such as CLIP-seq and CLASH studies. Two online tools, namely targetScan (version 7.1; http://www.targetscan.org/vert_71/),15 StarBase (version 2; http://starbase.sysu.edu.cn/starbase2/index.php)16 were used to explore variants located at the miRNA−messenger RNA binding site. The impact of variants on gene expression was evaluated by expression quantitative trait loci (eQTL) using the GTEx Portal (https://www.gtexportal.org/). RegulomeDB (https://regulomedb.org/) was applied to assess the potential regulatory functions of haplotype variants outside the coding region. The scores can range from 1 to 7, and lower scores are more likely to be related to regulatory variants. The range of probability is between 0 and 1, in which 1 denotes the most probability of being a regulatory variant.17 Also, the SNPInfo (SNP Function Prediction [FuncPred], https://snpinfo.niehs.nih.gov/) analysis was performed for all variants.18
7 RESULTS
7.1 COVID-19 associated genes
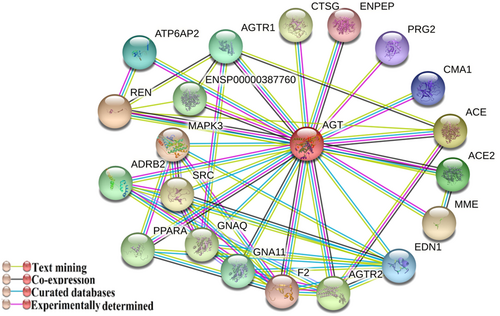
The literature review showed that angiotensin-converting enzymes (ACE and ACE2), as well as TMPRSS2, play main roles in COVID-19 infection. The protein−protein interaction analysis revealed the most important interacting genes associated with ACE. The results showed AGT (Angiotensinogen), AGTR2 (Angiotensin II Receptor Type 2), AGTR1 (Angiotensin II Receptor Type 1), KNG1 (Kininogen 1), REN (Renin), RHOA (Ras Homolog Family Member A), and RHOC (Ras Homolog Family Member C) were associated with ACE. The detailed results are shown in Figure 2A.

The AGT had a stronger association with ACE2 in protein-protein interactions, similar to protein−protein interactions in ACE (Figure 2A,B). The ACE2 and TMPRSS2 had strong association with each other (Figure 2B,C). The protein−protein interaction of AGT confirmed its associations with ACE2 and ACE (Figure 3). However, according to the interactions and also based on the pathway analysis, the results revealed that ACE, ACE-2, and AGT are involved in similar pathways, including the renin-angiotensin system (RAS) (KEGG, https://www.pharmgkb.org/pathway/PA165110622), and ACE Inhibitor pathways, (https://www.ncbi.nlm.nih.gov/biosystems/198763, https://www.pharmgkb.org/pathway/PA2023).
7.2 COVID-19 associated genes and variants involved in COVID-19 related traits
The GWA studies were mined to find variants of the candidate genes associated with COVID-19 related traits. ACE gene variants showed 10 unique polymorphisms, all of which were significantly associated with COVID-19 related traits including systolic and diastolic blood pressure, blood protein, and serum metabolite. Results are shown in Table 1. We also investigated GWAS variants located in the proximity of the genomic position of ACE. Two intragenic variants, rs4968782 and rs4277405, very close to the PPIAP55 pseudo gene, were associated with COVID-19 related traits (Table 2). No GWAS-associated variants were found in ACE2, and GWAS variants of TMPRSS2 were not associated with COVID-19 related traits (Table 3).
| Traits | Study population | Sample size | Variants | Type of variant | p Value | References |
|---|---|---|---|---|---|---|
| ACE activity | Han Chinese | 1023 | rs4343 | Synonymous | 3e-25 | 19 |
| Blood metabolite levels | European | 7824 | rs4351 | Intron | 4e-22, 1e-19, 3e-17, 1e-14 | 20 |
| 7824 | rs4362 | Synonymous | 1e-21 | 20 | ||
| Blood metabolite ratios | European | 7358 | rs4343 | Synonymous | 1e-37 | 20 |
| Blood protein levels | European | 3200 | rs4344 | Intron | 9e-136 | 21 |
| Diastolic blood pressure | European, Latino, Africa, East Asian, South Asian, mixed and unknown | 321262 | rs4295 | Intron | 4e-8 | 22 |
| European | 331204 | rs4308 | Intron | 7e-14 | 22 | |
| 378379 | rs4459609 | Intergenic | 2e-7,2e-14 | 23 | ||
| Metabolic traits | European | 2820 | rs4329 | Intron | 8e-20 | 24 |
| Metabolite levels | European | 891 | rs4351 | Intron | 9.00e-13 | 25 |
| Serum metabolite levels | European | 1678 | rs4325 | Intron | 1e-11, 3e-12 | 26 |
| African, European | 2938 | rs4343 | Synonymous | 9-25, 1e-18, 8e-14, 1e-16 | 26, 27 | |
| 1678 | rs4351 | Intron | 4e-14, 5e-15 | 26 | ||
| Systolic blood pressure | European, African, Hispanic, Asian, Native American | 776078 | rs4291 | Regulatory_region | 1e-9 | 28 |
| European, Latino, Africa, East Asian, South Asian, mixed and unknown | 321262 | rs4295 | Intron | 1e-8 | 22 |
| Traits | Study population | Sample size | Variants | Type of variant | p Value | References |
|---|---|---|---|---|---|---|
| CSF levels of AD proteins | Unknown | 574 | rs4968782 | Intragenic | 4e-12 | 29 |
| Diastolic blood pressure and alcohol interaction | European, African, Asian, Hispanic, or Latino American | 574163 | rs4277405 | Intragenic | 1e-12 | 30 |
| Diastolic blood pressure and smoking | European, African, Asian, Hispanic | 1191328 | rs4968782 | Intragenic | 3-e16, 3e-14, 7e-16, 4e-13 | 31 |
| Systolic blood pressure and alcohol interaction | European, African, Asian, Hispanic, or Latin American | 574117 | rs4277405 | Intragenic | 2e-12 | 30 |
| Systolic blood pressure and smoking status | European, African, Asian, Hispanic | 1191328 | rs4968782 | Intragenic | 8e-15, 2e-13, 6e-14, 3e-12 | 31 |
In addition, GWAS results were mined to identify genes having interactions with ACE. The results identified 39 GWAS variants. All KNG1 variants (12 variants), 6 polymorphisms in the AGT gene, 2 in the REN gene, 1 in the BDKRB2, and 1 in the RHOA gene were significantly associated with COVID-19 related traits. The results are shown in Table 4. Most of the associated traits were similar to those identified in association with ACE gene variants (Table 1). Polymorphisms within RHOC, BDKRB2, AGTR1, and AGTR2 genes were not related to COVID-19 related traits. GWAS variants of other COVID-19 related traits, including cold, respiratory diseases, and cardiovascular diseases, were assessed; however, no association was found for these traits.
| Genes | Traits or diseases | Study population | Sample size | Variants | Type of variant | p Value | References |
|---|---|---|---|---|---|---|---|
| REN | Systolic blood pressure | European | 422000 | rs72749713 | Intron | 9E-10 | 34 |
| Blood protein levels | European | 3301 | rs193280350 | Intron | 1E-15 | 35 | |
| Psychosis proneness | European | 3967 | rs7513165 | Intron | 3E-7, 2E-6 | 36 | |
| BDKRB2 | Mumps | European | 85380 | rs11160318 | Regulatory_region | 5E-12 | 37 |
| Coronary artery calcified atherosclerotic plaque in type 2 diabetes | African | 896 | rs7158214 | Regulatory_region | 4E-06 | 38 | |
| Subcutaneous adipose tissue | European | 2513 | rs4384548 | Regulatory_region | 5E-08 | 39 | |
| Corneal astigmatism | European | 86335 | rs56274409 | Intron | 7E-06 | 40 | |
| Cerebrospinal fluid AB1-42 levels | unknown | 3146 | rs76881547 | Regulatory_region | 5E-06 | 41 | |
| Gout | Han Chinese | 62 | rs2069590 | 3_prime_UTR | 1E-07 | 42 | |
| AGTR2 | Cystic fibrosis severity | European | 3476 | rs1403543 | Intron | 2E-06 | 43 |
| AGTR1 | Adolescent idiopathic scoliosis | Chinese | 499 | rs12695894 | Intron | 5E-12 | 44 |
| AGT | Diastolic blood pressure | European, Latino, African, African, East Asian, South Asian, mixed and unknown | 699641 | rs2004776 | Intron | 9E-7, 2E-11 | 22, 23 |
| Mean arterial pressure | European, African, Hispanic | 146562 | rs699 | Missense | 4E-12 | 45 | |
| Systolic blood pressure | European | 342415 | rs2493134 | Intron | 1E-12 | 46 | |
| Diastolic blood pressure | European | 342415 | rs2493134 | Intron | 1E-14 | 46 | |
| Coronary artery disease | unknown | 547261 | rs699 | Missense | 2E-08 | 47 | |
| Diastolic blood pressure and alcohol interaction | European, African, Asian, Hispanic, or Latin American | 535163 | rs2004776 | Intron | 9E-13 | 30 | |
| Diastolic blood pressure and smoking interaction | European, African, Asian, Hispanic | 610091 | rs2004776 | Intron | 2E-14 | 31 | |
| Systolic blood pressure and smoking interaction | European, African, Asian, Hispanic | 610091 | rs2004776 | Intron | 3E-10 | 31 | |
| Blood protein levels | European | 3200 | rs3789678 | Intron | 2E-16 | 21 | |
| Pancreatic ductal adenocarcinoma | European | 21536 | rs1326889 | Intron | 4E-07 | 48 | |
| Systolic blood pressure | European, African, Hispanic, Asian, Native American | 776078 | rs699 | Missense | 6E-26 | 28 | |
| Systolic blood pressure | European, Latino, African, African, East Asian, South Asian, mixed and unknown | 321262 | rs2004776 | Intron | 1E-09 | 22 | |
| Cardiovascular disease | European | 459000 | rs943580 | Intergenic | 4E-16 | 34 | |
| Systolic blood pressure | European | 422000 | rs2478539 | Intron | 3E-30 | 34 | |
| RHOC | Glomerular filtration rate (creatinine) | European | 110517 | rs12144044 | 5_prime_UTR | 3E-8 | 49 |
| Cerebral amyloid deposition in APOEe4 non-carriers | European and other | 370 | rs76570619 | Intron | 3E-7 | 50 | |
| RHOA | Educational attainment | European | 280007 | rs148734725 | Intron | 5E-12 | 51 |
| Cognitive ability | Intron | ||||||
| High intelligence | European | 87740 | rs1987628 | Intron | 4E-8 | 52 | |
| Morning person | European | 651295 | rs62259941 | Intron | 1E-17 | 53 | |
| Household income | European, unknown | 505541 | rs11716948 | Intron | 1E-10 | ||
| Intelligence | European | 269867 | rs7623659 | Intron | 1E-27 | 54 | |
| Educational attainment | European | 1131881 | rs116656374 | Intron | 3E-10 | 55 | |
| Cognitive performance | European | 402382 | rs7623659 | Intron | 4E-57 | 55 | |
| Coronary artery disease | European, unknown | 547261 340599 304591 |
rs7623687 | Intron | 1E-8, 4E-10, 2E-8 | 47, 56, 57 | |
| KNG1 | Activated partial thromboplastin time | European, Japanese | 377667 1431 9240 |
rs2304456 | Missense | 4E-45, 3E-199 2E-36 |
58-60 |
| Blood metabolite levels | European | 7824 | rs4686799 | Intron | 9E-6 | 20 | |
| Blood protein levels | European | 3301 997 3200 |
rs5030081 rs3856930 rs2304456 rs5030049 rs5030062 rs138610068 rs710446 rs5030044 |
Intron Intron Intron Intron Exon Intron Missense Intron |
4E-49 8E-15 3E-97 1E-96 6E-13 1E-13 2E-302 1E-300 |
21, 35, 61 | |
| Factor XI | European | 18214 | rs5030062 | Exon | 3E-40 | 62 | |
| Plasma renin activity | European | 5275 | rs5030062 | Exon | 2E-35 | 63 | |
| Small vessel stroke | Chinese | 3526 | rs1403694 | Intron | 1E-12 | 64 | |
| Thrombosis | European | 258962 | rs698078 | Intron | 5E-7 | 65 | |
| Venous thromboembolism | European | 187204 | rs710446 | Missense | 2E-203 | 66 |
7.3 Haplotype analysis of COVID-19 related traits
The haplotype analysis was performed based on 1000 Genomes Project haplotypes. First, all common variants in the ACE gene and its near genomic positions, related to blood metabolite levels, were analyzed (Tables 1 and 2). The results showed that all of these variants (rs4329_rs4325_rs4343_rs4351_rs4344_rs4362) were in strong linkage disequilibrium (LD) (r2 ≥ 0.90 and D′ ≥ 0.98). To examine the functional effect of these variants, the annotation of polymorphisms with eQTL was performed, and except for rs4343, all other variants were eQTL. The haplotype analysis was carried out on other variants associated with COVID-19 related traits. The results showed that rs4295_rs4968782_rs4308_rs4459609_rs4277405_rs4291 in the ACE gene and its near genomic positions (Tables 1 and 2) were in strong LD (r2 ≥ 0.88 and D′ ≥ 0.95). All these variants were eQTL and associated with blood pressure. Also, the haplotype analysis of AGT variants was conducted, and it was found that rs2478539_rs699_rs2493134_rs943580 were in a haplotype block (r2 ≥ 0.87 and D′ ≥ 0.96) for systolic blood pressure variations. All variants in this haplotype were also eQTL, showing the functional importance of this haplotype. Finally, the online tools targetScan and StarBase were used to find the miRNA binding site variants, while RegulomeDB and SNPinfo were applied to assess the potential regulatory functions of haplotype variants. The results are shown in Table 5. The RegulomeDB analysis indicated that except for three polymorphisms, other variants had a probability of 0.61≥ (ranked 4 or less) for the potential regulatory functions. The SNPinfo analysis showed that many of the index variants among identified haplotypes were located in the transcription factor binding sites (TFBS), exonic splicing enhancers (ESE), or silencer (ESS) activity variants. The three index polymorphisms (rs4968782, rs4277405, and rs4459609) had low probability scores in RegulomeDB and were TFBS variants based on SNPinfo.
| Haplotype | rsID | RegulomeDB | SNPinfo | eQTL | ||||
|---|---|---|---|---|---|---|---|---|
| Type of SNP | Probability | Ranking | TFBSa | Splicing (ESE or ESS)b | RegPotenti alc | |||
| ACE associated with blood metabolite variations | Index | rs4325 | 0.85 | 3a | NA | Yes | ||
| rs4329 | 0.61 | 4 | 0.13 | Yes | ||||
| rs4343 | 0.87 | 2b | Yes | 0 | - | |||
| rs4344 | 0.61 | 4 | 0.05 | Yes | ||||
| rs4351 | 0.61 | 4 | 0 | Yes | ||||
| rs4362 | 0.64 | 3a | Yes | 0.33 | Yes | |||
| Proxy | rs4316 | 0.83 | 2a | 0.37 | Yes | |||
| rs4320 | 0.61 | 4 | 0 | Yes | ||||
| rs4321 | 0.13 | 5 | 0.04 | Yes | ||||
| rs4323 | 0.13 | 5 | NA | Yes | ||||
| rs4324 | 0.59 | 5 | NA | Yes | ||||
| rs4326 | 0.61 | 4 | NA | Yes | ||||
| rs4327 | 0.61 | 4 | NA | Yes | ||||
| rs4330 | 0.61 | 4 | 0 | Yes | ||||
| rs4332 | 0.61 | 4 | 0.17 | Yes | ||||
| rs4333 | 0.61 | 4 | 0.11 | Yes | ||||
| rs4334 | 0.13 | 5 | 0 | Yes | ||||
| rs4335 | 0.13 | 5 | 0 | Yes | ||||
| rs200491776 | 0.13 | 5 | - | |||||
| rs4336 | 0.13 | 5 | 0.18 | Yes | ||||
| rs201390711 | 0.13 | 5 | - | |||||
| rs4337 | 0.13 | 5 | 0.20 | Yes | ||||
| rs1987692 | 0.61 | 4 | NA | Yes | ||||
| rs4341 | 0.61 | 4 | 0 | - | ||||
| rs4342 | 0.61 | 4 | 0 | - | ||||
| rs4353 | 0.74 | 2b | 0 | Yes | ||||
| rs4359 | 0.78 | 3a | 0 | Yes | ||||
| rs4363 | 0.61 | 4 | 0.59 | Yes | ||||
| rs1055086 | 0.61 | 4 | 0 | Yes | ||||
| ACE haplotype associated with blood pressure variations | Index | rs4968782 | 0.18 | 7 | Yes | 0 | Yes | |
| rs4277405 | 0.13 | 5 | Yes | 0 | Yes | |||
| rs4459609 | 0.13 | 5 | Yes | 0.12 | Yes | |||
| rs4291 | 0.61 | 4 | Yes | 0.42 | Yes | |||
| rs4295 | 0.61 | 4 | 0.12 | Yes | ||||
| rs4308 | 0.61 | 4 | 0 | Yes | ||||
| Proxy | rs6504163 | 0.36 | 3a | NA | Yes | |||
| rs8077276 | 0.13 | 5 | NA | Yes | ||||
| rs4968783 | 0.59 | 5 | NA | Yes | ||||
| rs4292 | 0.70 | 4 | Yes | 059 | Yes | |||
| AGT haplotype associated with systolic blood pressure variations | Index | rs943580 | 0.61 | 4 | 0 | Yes | ||
| rs2478539 | 0.61 | 4 | 0 | Yes | ||||
| rs699 | 0.61 | 4 | Yes | 0.46 | Yes | |||
| rs2493134 | 0.61 | 4 | Yes | 0 | Yes | |||
| Proxy | rs2493126 | 0.61 | 4 | 0.19 | Yes | |||
| rs2493128 | 0 | 5 | 0 | Yes | ||||
| rs2006765 | 0.61 | 4 | NA | Yes | ||||
| rs2493133 | 0.79 | 2b | 0.10 | Yes | ||||
| rs2478543 | 0.79 | 3a | 0.07 | Yes | ||||
| rs5051 | 1.0 | 2b | 0.23 | Yes | ||||
| rs2493135 | 0.61 | 4 | NA | Yes | ||||
| rs2493136 | 0.61 | 4 | NA | Yes | ||||
- Note: Bold values shows proxy variants with more regulatory effects. Ranking/supporting data: 2a/TF binding + matched TF motif + matched DNase Footprint + DNase peak; 2b/TF binding + any motif + DNase Footprint + DNase peak; 3a/TF binding + any motif + DNase peak; 4/TF binding + DNase peak; 5/TF binding or DNase peak; 7/Other.
- a affecting the transcription factor binding sites (TFBS) activity (transcriptional regulation);
- b exonic splicing enhancers (ESE) or silencers (ESS);
- c Regulatory Potential Score only for SNPs that are outside of the coding region in SNP selection.
The haplotype analysis showed proxy haplotypic variants were associated with described traits. Most of the proxy haplotype variants identified form the GWA studies were eQTL. Three variants, including rs4316, rs4353, and rs4359, and three variants rs2493133, rs2478543, and rs5051, were correlated with blood metabolite levels and systolic blood pressure, respectively. These variants showed more regulatory effects compared with other GWAS variants, suggesting the potential effects of these variants on blood metabolite and blood pressure.
8 DISCUSSION
Genetic factors play a key role in the susceptibility and severity of COVID-19. In this study, we tried to identify variants and haplotypes associated with COVID-19 related traits by the identification of genes and GWAS variants associated with COVID-19. The literature review showed that ACE, ACE2, and TMPRSS2 genes play fundamental roles in COVID-19 infection.67 The results showed GWAS variants in ACE and AGT genes were the most significant variants associated with COVID-19 related traits. These variants contribute to the development of blood pressure and blood metabolite levels and are potentially involved in COVID-19 severity. Two specific haplotypes were found in the ACE gene, closely related to blood metabolites levels and blood pressure. GWAS index variants of these haplotypes functionally affect the ACE expression. Three candidate proxy variants, including rs4316, rs4353, and rs4359, were identified to be associated with blood metabolite levels. The effect of these variants on blood metabolites levels was not assessed in previous studies. Studies have shown that the ACE-2 and ACE genes are correlated with COVID-19 infection.1, 2, 6, 7, 68, 69 For example, infected individuals with COVID-19 receiving ACE inhibitors or the angiotensin receptor blocker (ACEis/ARBs) had a lower risk of mortality than other COVID-19 patients.68 Several studies have been performed on the role of ACE inhibitors in the development of blood pressure or blood metabolic levels,70-74 confirming the results obtained in this study. Also, a relationship between blood pressure control and its role in the mortality of COVID-19 patients has been observed, implying the role of systolic and diastolic blood pressure controlling drugs in COVID-19 infection.68 Another study examined ACE gene haplotypes in the African American population and their close association with blood pressure. The study identified rs4313-rs4337 haplotype in association with hypertension.75 The relationship between blood metabolites and ACE gene polymorphisms has been investigated in previous studies. Also, the correlation of ACE I/D polymorphisms with nitric oxide metabolite and blood pressure levels in Mexican men has been identified.76 A recent study carried out on the effect of the ACE gene on mortality rate in COVID-19 shows that in the European population, ACE Del/Del polymorphism is significantly involved in COVID-19-associated mortality. The authors suggested further research on this variant and level of ACE as prognostic biomarkers for the severity of COVID-19. They have identified both ACE and Ang II could act as potential targets for the treatment of COVID-19.6 In addition, recent studies identified variants in the ACE gene, which are implicated in COVID-19 outcome.7 Recently, a pilot study suggested that the rs4343 variant in ACE may be a predictive marker for the severity of COVID-19 in patients with hypertension.5 Also, this variant is associated with angiotensin-converting enzyme inhibitors.77 In addition, rs4343, rs4329, and rs4351 are associated with blood metabolic measurements.20, 24, 25 A recent study showed the role of rs699 in the prediction of the clinical outcome of SARS-CoV-2 infection.78 These findings confirm the importance of variants identified as haplotypes in the current study. Also, the ACE2 gene was assessed as the receptor of COVID-19.79, 80 It was revealed that its polymorphisms are associated with the genetic susceptibility of COVID-19.81 No GWAS-associated variants were found to be related to ACE2.
In this study, ACE and AGT variants had associations with blood pressure. As shown in Figures 2 and 3, AGT is closely associated with both COVID-19 related genes (ACE, ACE2), which is in agreement with the previous study.3 The study identified a haplotype and three rs2493133, rs2478543, and rs5051 variants in the AGT gene associated with systolic blood pressure. The role of rs2493133 and rs2478543 variants in blood pressure was not evaluated in previous studies, and the results of previous studies on the role of rs5051 in hypertension were not similar.82-84 The role of AGT gene polymorphisms in blood pressure was investigated in previous studies.85-87 Polymorphisms in this gene play a role in respiratory distress syndrome.88 This gene is co-expressed with renin in the brain.89 AGT, ACE, and ACE2 are related to vasoconstriction, vasodilation, and angiogenesis, as well as the risk of developing ischemic stroke. The ACE2-Ang-(1-7)-Mas axis lowers the ACE-Ang II-AT1R axis and, accordingly, exerts a protective effect against stroke and shows anti-hypertensive properties.3 However, there was no study on the association of AGT levels with the severity of COVID-19.90 The AGT protein is a potential therapeutic target to decrease infection.4 Recent studies found an association between the expression of AGT and ACE2 expression.4 Both of these genes are involved in RAS.2 The inhibition of this system may be a therapeutic option and an important pathway for COVID-19 treatment.1, 91
9 CONCLUSION
According to the results, all three index variants in candidate haplotypes had important effects on blood pressure and blood metabolite levels. The findings were completely specific, and variants are effective in gene expression. Furthermore, three candidate variants, rs4316, rs4353, and rs4359, and three candidate variants, rs2493133, rs2478543, and rs5051 were identified to be involved in blood metabolite levels and systolic blood pressure, respectively. More observational studies on identified variants are needed to confirm the association of these candidate variants with blood metabolite levels, blood pressure, and COVID-19 severity.
AUTHOR CONTRIBUTIONS
All authors contributed to the conception and design of the study. The literature review and pathway analysis were performed by Mandana Hasanzad and Bagher Larijani. GWAS variants associated with COVID-19-related traits were identified by Marziyeh Zoughi and Morteza Gholami. Haplotype and functional analyses were conducted by Mahsa M. Amoli and Morteza Gholami. The manuscript was drafted by Morteza Gholami and Marziyeh Zoughi with the help of Mahsa M. Amoli. All authors commented on previous versions of the manuscript and discussed the results. Mahsa M. Amoli supervised the project and approved the final version. All authors read and approved the final version of the manuscript.
ACKNOWLEDGMENT
This study was supported by the Tehran University of Medical Sciences (Grant No. 51229-221-3-99).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ETHICS STATEMENT
This study was approved by the Research Ethics Committee of Tehran University of Medical Sciences.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




