Nonpharmaceutical interventions for COVID-19 disrupt the dynamic balance between influenza A virus and human immunity
Qing Ye and Huihui Liu contributed equally to this study.
Abstract
During the COVID-19 epidemic, nonpharmaceutical interventions (NPIs) blocked the transmission route of respiratory diseases. This study aimed to investigate the impact of NPIs on the influenza A virus (IAV) outbreak. The present study enrolled all children with respiratory tract infections who came to the Children's Hospital of Zhejiang University between January 2019 and July 2022. A direct immunofluorescence assay kit detected IAV. Virus isolation and Sanger sequencing were performed. From June to July 2022, in Hangzhou, China, the positive rate of IAV infection in children has increased rapidly, reaching 30.41%, and children over 3 years old are the main infected population, accounting for 75% of the total number of infected children. Influenza A (H3N2) viruses are representative strains during this period. In this outbreak, H3N2 was isolated from a cluster of its own and is highly homologous with A/South_Dakota/22/2022 (2021–2022 Northern Hemisphere). Between isolated influenza A (H3N2) viruses and A/South_Dakota/22/2022, the nucleotide homology of the HA gene ranged from 97.3% to 97.5%; the amino acid homology was 97%–97.2%, and the genetic distance of nucleotides ranged from 0.05 to 0.052. Compared with A/South_Dakota/22/2022, the isolated H3N2 showed S156H, N159Y, I160T, D186S, S198P, I48T, S53D, and K171N mutations. There was no variation in 13 key amino acid sites associated with neuraminidase inhibitor resistance in NA protein. Long-term NPIs have significantly affected the evolution and transmission of the influenza virus and human immunity, breaking the dynamic balance between the IAV and human immunity.
1 INTRODUCTION
Influenza is an acute respiratory infectious disease caused by the influenza virus. It often causes worldwide pandemics or local small and medium-sized epidemics.1-3 The Lancet recently pointed out in 2018 that there are an estimated 1 billion cases of influenza a year, of which 3–5 million are severe, causing 290 000–650 000 deaths.4 To avoid pre-existing immunity and gain a competitive advantage, the influenza virus has a high mutation ability through the mutation of surface proteins in the evolutionary process.2, 5 Based on changes in glycoproteins, hemagglutinin (HA), and neuraminidase (NA), many subtypes of influenza A virus (IAV) have been found.6 H3N2 has become the dominant strain of influenza in most Asian countries due to its rapid antigenic drift.7 In China, 7 of the 10 minor epidemics in 1968–1992 were caused by the H3N2 subtype, and an extensive range of H3N2 influenza epidemics occurred in 1998 and 2002.8 During the COVID-19 outbreak, H3N2 predominated in many countries. Genetically, since September 2020, influenza A (H3N2) viruses belong to 3C.2alb.
The global persistence of H3N2 IAV results from a migrating metapopulation in which multiple different localities may seed seasonal epidemics in temperate regions in a given year.9-12 Therefore, population density and regional connectivity are essential in maintaining the virus population. The rapid and extensive global circulation provides many opportunities for antigen selection of influenza viruses. However, the emergence of COVID-19 has led to great changes in people's lifestyles, and nonpharmaceutical interventions (NPIs) have effectively cut off the transmission chain. Since the emergence of COVID-19, China has implemented very strict NPIs, including hand hygiene, use of face masks, social distance, limiting mass gatherings, school closures, and standing at home orders. Therefore, there are significant differences in people's immunity to each influenza virus. Due to the fluctuation of the population level of immune pressure, the speed of antigen selection of IAV lineages changes with time.11, 13 The outbreak of the COVID-19 pandemic in 2020 seriously affected the genetic and antigenic diversity of IAVs.
The human body forms specific immune barriers mainly through infection and vaccination. However, the emergence of COVID-19 and the implementation of NPIs effectively cut off the respiratory transmission chain. The human body cannot obtain immunity by infecting the influenza virus. Long-term suppression of virus transmission will reduce population immunity, thereby increasing the severity of future influenza virus epidemics.12, 14 Therefore, by monitoring IAV, this study explored the epidemic characteristics of IAV before and after the outbreak of COVID-19 and the impact of NPIs on the IAV epidemic.
2 METHODS
2.1 Patient selection
The present study enrolled all children with respiratory tract infections who came to the Children's Hospital of Zhejiang University between January 2019 and July 2022 to explore the variations in the prevalence of IAV in children in Hangzhou, China. Based on the criteria of the U.S. Centers for Disease Control and Prevention, the enrolled children met the following criteria: (1) children aged younger than 18 years; (2) confirmed fever (a body temperature above 37.5°C); and (3) one or more respiratory symptoms within 14 days of onset (cough, sore throat, sputum, shortness of breath, lung auscultation abnormality (rale or wheeze), tachypnea, and chest pain). The exclusion criteria for this study were (1) repeated visits within a week; (2) hospital infection; (3) congenital pulmonary airway malformation and an impaired immune system; and (4) COVID-19 infection. All the enrolled children were divided into five age groups: under 28 days of age (0–28 d), 1–12 months of age (>1–12 mo), 1–3 years of age (>1–3 y), 3–6 years of age (>3–6 y), and more than 6 years of age (>6 y).
2.2 Data and specimen collection
Demographic characteristics from enrolled children were obtained from their electronic medical records. Respiratory specimens (nasopharyngeal aspirates/bronchoalveolar lavage fluid) were obtained from all the enrolled children as soon as they were admitted by trained staff following standard operating procedures. The specimens were immediately transferred to the clinical laboratory. The Respiratory Virus Antigen Detection Kit (Genesis) was used to detect IAV.
2.3 Molecular identification of influenza virus
The primers for HA and NA gene amplification of influenza A (H3N2) viruses were designed by Primer Premier 5.0 software: HA (F: 5′-AGARATCATTGCTTGAGC-3′, R: 5′-GCCTGAAAACCGTACCAAAC-3′), NA (F: 5′-GCAGGAGAGTAATGAATGAATC-3′; R: 5′-TGCGAAAGCTATGATATGGC-3′); the lengths of the amplified products were 1107 and 1431 bp, respectively. A PrimeScript One-Step RT amplified the HA and NA genes‒PCR Kit (Takara, No. RR055A) using genomic RNA of the H3N2 subtype as a template. The reaction system was prepared according to the instructions of the kit. The amplification conditions were as follows: 50°C, 30 min; 94°C 2 min; 94°C 30 s, 50°C 40 s, 72°C 90 s, 30 cycles; 72°C 11 min. The amplified DNA products were confirmed by 1.5% agarose gel electrophoresis and purified and recovered.
Log into the National Biotechnology Information Center and download the A/South_Dakota/22/2022 (2021–2022 Northern Hemisphere) and some representative strains of influenza A (H3N2) viruses worldwide were used as reference strains. Sanger sequencing was used to determine the whole genome sequences of the influenza A (H3N2) isolated viruses. Mega 7.0.14 was used for sequence alignment, and neighbor-joining was used to construct the HA gene phylogenetic tree by comparison with the reference strain. The HA gene sequence of isolated viruses was analyzed by MCMC with BEAST1.8.4.
2.4 Statistical analysis
Categorical variables are presented as counts with percentages. Proportions for categorical variables were compared using the chi-square test or Fisher's exact test. A value of p < 0.05 was considered statistically significant. Statistical analyses were conducted in SPSS version 26.0 software (IBM).
3 RESULTS
3.1 Incidence of IAV infection between January 2019 and July 2022
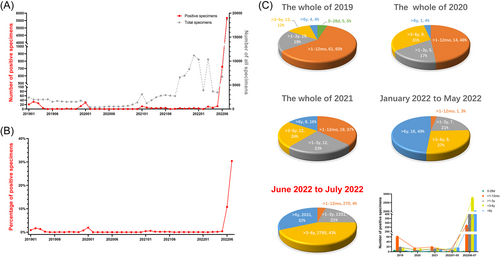
A total of 109 990 specimens in our hospital were detected for IAV between January 2019 and July 2022. The detailed demographic characteristics are shown in Table 1. The number and percentage of IAV-positive specimens are depicted in Figures 1A,B. From January 2019 to December 2021, the prevalence of IAV in children peaked only in winter and remained at a low level the rest of the time. However, since 2022, the number of children with IAV infection has shown explosive growth. In June and July 2022, the number of IAV-infected children was 724 and 5674, respectively, and the detection rates were 10.75% and 30.41%, respectively. In addition, from 2019 to 2021, children under 1 year old were the main population with IAV infection, accounting for 65%–37% of the total number of infected children. However, from 2020, in the proportion of all age groups in the total number of IAV infections, the ratio of children under 1 year old gradually decreased, and the ratio of children over 3 years old gradually increased. In the last 2 months, 75% of the children with IAV infection were children over 3 years old, while only 4% were children under 1 year old (Figure 1C). There was no significant difference in sex among IAV-infected children (p > 0.05).
| 2019 (n = 21 452) | 2020 (n = 8202) | 2021 (n = 52 879) | January 2022 to May 2022 (n = 31 716) | June 2022 to July 2022 (n = 25 395) | |
|---|---|---|---|---|---|
| IAV | 101/21 452 (0.47) | 29/8202 (0.35) | 51/52 879 (0.10) | 33/31 716 (0.10) | 6398/25 395 (25.19) |
| Age | |||||
| 0–28 days | 5/2335 (0.21) | 0/943 (0) | 0/222 (0) | 0/77 (0) | 1/22 (4.55) |
| >1–12 months | 61/7010 (0.87) | 14/2825 (0.50) | 19/5084 (0.37) | 1/2163 (0.05) | 104/777 (16.06) |
| >1–3 years | 19/5053 (0.38) | 5/1985 (0.25) | 12/14 968 (0.08) | 7/6634 (0.11) | 521/3210 (19.53) |
| >3–6 years | 12/4283 (0.28) | 9/1603 (0.56) | 12/18 015 (0.07) | 9/10 491 (0.09) | 1160/5636 (26.84) |
| >6 years | 4/2771 (0.14) | 1/846 (0.12) | 8/14 590 (0.05) | 16/12 351 (0.13) | 789/3408 (30.88) |
| Gender | |||||
| Male | 69/12 191 (0.57) | 19/4701 (0.40) | 32/5210 (0.61) | 19/16 906 (0.11) | 3616/14 128 (25.59) |
| Female | 32/9261 (0.35) | 10/3501 (0.29) | 19/24 408 (0.08) | 14/14 810 (0.09) | 2782/11 267 (24.69) |
- Note: Data were expressed as the IAV positive number/the total number (%).
- Abbreviation: IAV, influenza A virus.
3.2 Phylogenetic analysis of H3N2 viruses
Considering the high detection rate of IAV infection in children, we randomly selected 320 IAV-positive samples from June to July 2022 according to a proportion of 5%. Based on RT-PCR, influenza A (H3N2) lineages are the representative strain during this outbreak. Then, five isolated influenza A (H3N2) virus strains were selected for whole genome sequencing.
3.2.1 Homology analysis
By MegAlign in DNASTAR software, the determined sequence of influenza A (H3N2) viruses was compared with A/South_Dakota/22/2022 (2021–2022 Northern Hemisphere) to analyze nucleotide and amino acid homology. As shown in Table 2, in five isolated influenza A (H3N2) virus strains, the nucleotide homology range of the HA gene was 99.4%–99.7%, and the amino acid homology range was 99.1%–100%. Compared with A/South_Dakota/22/2022, the nucleotide homology of the HA gene ranged from 97.3% to 97.5%; the amino acid homology was 97%–97.2%. The average genetic distances of the five influenza A (H3N2) virus strains were isolated and analyzed using the Kimura 2-parameter model in MEGA software. As shown in Table 3, the nucleotide genetic distance of the five influenza A (H3N2) virus strains ranged from 0 to 0.01. Their genetic distance to A/South_Dakota/22/2022 ranged from 0.025 to 0.028.
| Specimens | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| 1. ZXH_Segment4 | *** | 99.5 | 99.6 | 99.6 | 99.5 | 97.5 |
| 2. SYH_Segment4 | 99.6 | *** | 99.5 | 99.6 | 99.6 | 97.5 |
| 3. WYC_Segment4 | 99.5 | 99.1 | *** | 99.4 | 99.5 | 97.5 |
| 4. HJH_Segment4 | 99.8 | 99.5 | 99.3 | *** | 99.7 | 97.3 |
| 5. CCT_Segment4 | 100 | 99.6 | 99.5 | 99.8 | *** | 97.3 |
| 6. A/South_Dakota/22/2022 (2021-2022_Northern_Hemispher) | 97.2 | 97.2 | 97 | 97 | 97.2 | *** |
- ***Indicates that the same sequence does not need to be compared.
| Specimens | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| 1. ZXH_Segment4 | ||||||
| 2. SYH_Segment4 | 0.01 | |||||
| 3. WYC_Segment4 | 0.00 | 0.01 | ||||
| 4. HJH_Segment4 | 0.00 | 0.00 | 0.006 | |||
| 5. CCT_Segment4 | 0.01 | 0.00 | 0.005 | 0.003 | ||
| 6. A/South_Dakota/22/2022 (2021-2022_Northern_Hemispher) | 0.026 | 0.026 | 0.025 | 0.028 | 0.027 |
3.2.2 Genetic evolution analysis
The HA gene sequences of five influenza A (H3N2) virus strains were analyzed and compared with those of other regional or international representative strains published by NCBI. The HA gene evolutionary tree (Figure 2A) was constructed by the Kimura 2-parameter model of the neighbor-joining method in MEGA software. The results showed that all five isolates belonged to 3C.2a1b, with a bootstrap value of 98. The five isolates formed a cluster (bootstrap value = 100) and clustered with A/South_Dakota/22/2022 into one branch (bootstrap value = 98). In addition, the five isolates were closely related to A/India/Pun-1920970/2019, A/Italy/9061/2019, A/Japan/8002/2019, and A/California/55/2020. It is speculated that the influenza A (H3N2) virus in Hangzhou recently evolved from them (Figure 2B).

To understand the evolution of influenza viruses, we constructed a data set of the HA gene of H3N2 between 2018 and 2022. First, the root-to-tip divergence regression analysis showed that H3N2 had obvious temporal evolution characteristics, and the R2 value reached 0.852. The evolution rate of the HA gene was 4.048*103/locus/year (95% confidence interval: 3.191*103 to 4.972*103). The evolution of the HA gene was close to a straight line before 2017, basically in a stable state. Since 2018, there has been a small peak, which means that it has a certain genetic variation in that year. From 2018 to 2020, the fluctuation was large, indicating great gene variations in viruses under different factors in these 2 years. From 2021 to 2022, it approaches a straight line and returns to a stable state (Figure 2C,D).
3.2.3 Antigenicity analysis
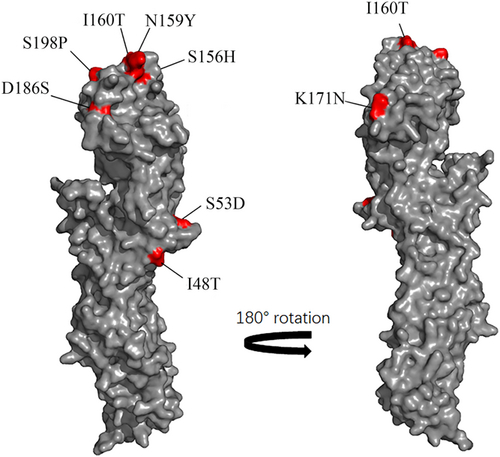
The epitopes of influenza A (H3N2) viruses were mainly located in the HA1 protein. The 131 amino acid position on the five antigenic sites (A–E) of the globular head of HA is the main target of specific antibodies. Antigenic drift is caused by the accumulation of amino acid substitutions at these sites.15, 16 Compared with A/South_Dakota/22/2022, five influenza A (H3N2) virus strains isolated had amino acid substitutions at three antigenic sites. The antigenic epitope mutations mainly occurred at positions 156, 159, 160, 186, and 198 at site B, 48, and 53 at site C, 171 at site D, and no mutations were detected at sites A and E (Table 4). All strains showed S156H, N159Y, I160T, D186S, S198P, I48T, S53D, and K171N mutations (Figure 3).
| Strains | A | B | C | D | E | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 138 | 142 | 144 | 145 | 128 | 156 | 159 | 160 | 186 | 198 | 45 | 48 | 50 | 53 | 278 | 280 | 311 | 312 | 171 | 212 | 214 | 219 | 230 | 246 | 62 | 75 | 78 | 94 | 260 | 261 | |
| A/South_Dakota/22/2022 (2021-2022_Northern_Hemispher) | A | G | S | S | T | S | N | I | D | S | N | I | E | S | K | E | Q | S | K | A | I | S | I | N | G | Q | G | N | I | R |
| ZXH_Segment4 | A | G | S | S | T | H | Y | T | S | P | N | T | E | D | K | E | Q | S | N | A | I | S | I | N | G | Q | G | N | I | R |
| SYH_Segment4 | A | G | S | S | T | H | Y | T | S | P | N | T | E | D | K | E | Q | S | N | A | I | S | I | N | G | Q | G | N | I | R |
| WYC_Segment4 | A | G | S | S | T | H | Y | T | S | P | N | T | E | D | K | E | Q | S | N | A | I | S | I | N | G | Q | G | N | I | R |
| HJH_Segment4 | A | G | S | S | T | H | Y | T | S | P | N | T | E | D | K | E | Q | S | N | A | I | S | I | N | G | Q | G | N | I | R |
| CCT_Segment4 | A | G | S | S | T | H | Y | T | S | P | N | T | E | D | K | E | Q | S | N | A | I | S | I | N | G | Q | G | N | I | R |
3.2.4 Drug resistance analysis
NA protein is responsible for destroying sialic acid residues on the cell surface glycoprotein or glycolipid end so that the virus can be released from the host cell and spread further. Similar to HA protein, NA protein is another surface antigen of IAV. Currently, neuraminidase inhibitors (oseltamivir and zanamivir) targeting NA protein have become specific drugs for treating influenza. Compared with A/South_Dakota/22/2022, 13 key amino acid sites related to neuraminidase inhibitor resistance in five isolated influenza A (H3N2) virus strains did not change (Table 5).
| Strains | 119 | 136 | 151 | 222 | 224 | 245 | 246 | 247 | 248 | 276 | 292 | 294 | 371 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A/South Dakota/22/2022(2021-2022 Northern Hemisphere) | E | Q | D | I | R | N | A | T | G | E | R | N | R |
| Zhuxiheng Segment6 | E | Q | D | I | R | N | A | T | G | E | R | N | R |
| Sunyuhang Segment6 | E | Q | D | I | R | N | A | T | G | E | R | N | R |
| Huangjinghan Segment6 | E | Q | D | I | R | N | A | T | G | E | R | N | R |
| Wangyichen NA.seq | E | Q | D | I | R | N | A | T | G | E | R | N | R |
| Chenchutong NA.seq | E | Q | D | I | R | N | A | T | G | E | R | N | R |
4 DISCUSSION
Severe acute respiratory syndrome coronavirus 2 is a severe infectious respiratory virus that causes COVID-19 and damages almost all human organs, including the lungs, heart, kidneys, liver, and brain, even leading to death in severe cases.17-26 To contain the sudden COVID-19 epidemic, almost all countries in the world have not only recommended that their citizens be vaccinated against COVID-1927-34 but also adopted a series of strict NPIs, such as hand hygiene, use of face masks, social distance, limiting mass gatherings, school closures, and staying at home orders. NPIs disrupt the transmission chain of respiratory diseases, so they are equally effective in controlling other respiratory virus infections.35 Our previous studies found that the total number of children with respiratory symptoms in our hospital decreased by 61.77% in 2020 compared with that in 2019, and the number of children with common respiratory virus infection decreased by 64.59% compared with that in 2019,36-39 including atypical bacteria.36, 40 IAV was not detected from February 2020.38 Surprisingly, there are also effects on infections with intestinal pathogens and diseases such as allergies.41, 42 Although a wide range of NPIs interfered with the spread of the influenza virus from 2020 to 2021, the number of children infected with IAV has increased dramatically since January 2022, and the detection rate of IAV in July reached 30.41%. We speculate that the high infection rate of IAV may be due to the following three reasons. The first is the evolution of the virus, the second is the reduction of human immunity, and the last is the recovery of transmission routes.
Large-scale COVID-19 vaccination and strict long-term NPIs slow the global recovery of influenza, delay competition among existing influenza lineages, and further differentiate the spatially separated influenza lineage. However, it cannot be ignored that these individual influenza lineages will eventually expand, compete, and spread more widely once again.28 Recombination, mutation, and genome evolution among influenza viruses are some ways to increase the diversity of viral protein sequences, escape antibody pressure and escape the host immune system. To determine whether the evolution of the influenza lineage causes the high positive rate of IAV, we isolated and identified the virus. In this study, A (H3N2) lineages represent Hangzhou's strain during the last 2 months. Through further phylogenetic analysis of influenza A (H3N2) viruses, we found that A (H3N2) viruses isolated in this outbreak form a cluster of their own (the nucleotide homology of the HA gene ranged from 99.4% - 99.7%, the amino acid homology ranged from 99.1% to 100%, and the nucleotide genetic distance ranged from 0 to 0.01). In addition, it is also highly homologous with A/South_Dakota/22/2022 (the nucleotide homology of the HA gene ranged from 97.3% to 97.5%; the amino acid homology ranged from 97% to 97.2%, and the genetic distance of nucleotides ranged from 0.05 to 0.052).
However, it cannot be ignored that IAV is the most frequently mutated influenza virus, and amino acid substitutions at antigenic sites are able to cause antigenic drift, which increases the risk of influenza epidemics. It is generally believed that if more than four sites mutate in the antigenic determinants and are distributed on more than two antigenic determinants, a new antigenic strain appears.43 In the current study, five isolated influenza A (H3N2) virus strains had the same eight amino acid substitution sites, involving three antigenic determinants. In addition, a recent study showed that only seven amino acids around the HA receptor binding site of influenza A (H3N2) viruses were the main determinants of antigen change during 1968–2003, including 145, 155, 156, 158, 159, 189, and 193 of H3N2.44 A single amino acid change in one of the seven sites in the HA gene is sufficient to produce a new antigenic variation. Since 2003, the antigenic variation of influenza A (H3N2) viruses has been associated with these seven sites.45 In our study, substitutions S156H and N159Y were found in two of these key positions. Therefore, although influenza A (H3N2) viruses isolated in this outbreak are highly homologous with A/South_Dakota/22/2022, their receptor binding sites are different, suggesting that the antigenicity of influenza A (H3N2) viruses may have changed to some extent.
Human immunity is the most important barrier to protecting humans from the influenza virus. Lack of exposure to influenza will reduce population immunity and increase the severity of influenza in the event of a new influenza pandemic. During the COVID-19 epidemic, long-term isolation at home and wearing masks and other NPIs limited people's exposure to the influenza virus. Over time, immunity to the influenza virus declines in all age groups.14 In addition, the continuous evolution of the influenza virus increases the risk that the antigens in the influenza vaccine cannot represent the virus eventually transmitted, thus reducing the effectiveness of the influenza vaccine.46 According to the difference in antigenic sites of influenza A (H3N2) virus in this outbreak, the antigenicity of influenza A (H3N2) viruses may have changed to some extent. In this context, the protection for the 2021–2022 Northern Hemisphere vaccine strains recommended by the WHO is fraught with uncertainties. Further studies are needed to verify the protective effect of the vaccine. The present study also found that the 13 key amino acid sites associated with neuraminidase inhibitor resistance were not changed. This may suggest that neuraminidase inhibitors (oseltamivir and zanamivir) could still effectively treat the infection of influenza A (H3N2) viruses in this outbreak in Hangzhou, China.
With the gradual control of the COVID-19 pandemic and large-scale vaccination, the use of NPIs to limit transmission will gradually decrease. Global contacts slowly returned to pre-COVID-19 pandemic levels. These factors provide opportunities for the spread and evolution of influenza viruses. Recent studies have shown a correlation between the spread of influenza at the national level and the severity of the COVID-19 pandemic. These studies have come from countries with no influenza activity since the onset of the COVID-19 pandemic.47-50 In Hangzhou, China, to prevent the COVID-19 pandemic, the government has completed more than 30 million doses of vaccination, the basic immunization coverage rate is more than 90%, and the enhanced vaccination rate is more than 70%. The limitation of NPIs has been gradually reduced, and social activities have gradually returned to the normal level. This makes it possible for the influenza virus to spread from person to person. At the same time, children over 3 years old are in kindergartens or schools, and they are in close contact with each other. If there are cases of influenza virus infection, it is very easy to cause transmission. This explains why, from January 2022, children over 3 years old have become the main population of IAV infection.
Influenza surveillance is a key measure to prevent and control and an essential basis for the early prediction and early warning of influenza. From surveillance data for influenza A (H3N2) viruses in 2020 and 2021, we can see that it continues to spread in East Asia, South Asia, and Southeast Asia.28 The present study has continuously monitored its incidence in children in Hangzhou since January 2019. Long-term NPIs significantly affect the evolution and transmission of the virus as well as human immunity. Therefore, these measures may play an important role in the high prevalence of IAV infection. Considering the epidemic characteristics of influenza A (H3N2) viruses during the COVID-19 epidemic, attention should be given to the spread of other infectious diseases when eliminating the COVID-19 epidemic.
5 CONCLUSION
Long-term NPIs significantly affected the transmission and evolution of influenza viruses and population immunity, breaking the dynamic balance between influenza A and human immunity.
AUTHOR CONTRIBUTIONS
Qing Ye drafted the initial manuscript and contributed to manuscript editing. Qing Ye, Huihui Liu, Jianhua Mao, and Qiang Shu collected the data from patients and contributed to manuscript editing. Jianhua Mao and Qiang Shu devised the conceptual ideas, contributed to the discussion and interpretation of the results, and reviewed the final manuscript. All authors approved the final manuscript.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (Grant/Award number: 82270741), the Natural Science Foundation of Zhejiang Province (LY22H050001), the Key Project of Provincial Ministry Construction, Health Science and Technology Project Plan of Zhejiang Province (WKJ-ZJ-2128), and Key Research and Development Plan of Zhejiang Province (Grant/Award number: 2021C03079).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ETHICS STATEMENT
The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The Ethics Committee approved this study of the Children's Hospital, Zhejiang University School of Medicine (2021-IRB-228).
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request. All requests should be submitted to the corresponding author for consideration.




