GSK3326595 is a promising drug to prevent SARS-CoV-2 Omicron and other variants infection by inhibiting ACE2-R671 di-methylation
Zhongwei Li, Hongmei Yong, Wenwen Wang, and Yue Gao contributed equally to this study.
Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) caused COVID-19 epidemic is worsening. Binding of the Spike1 protein of SARS-CoV-2 with the angiotensin-converting enzyme 2 (ACE2) receptor mediates entry of the virus into host cells. Many reports show that protein arginine methylation by protein arginine methyltransferases (PRMTs) is important for the functions of these proteins, but it remains unclear whether ACE2 is methylated by PRMTs. Here, we show that PRMT5 catalyses ACE2 symmetric dimethylation at residue R671 (meR671-ACE2). We indicate that PRMT5-mediated meR671-ACE2 promotes SARS-CoV-2 receptor-binding domain (RBD) binding with ACE2 probably by enhancing ACE2 N-glycosylation modification. We also reveal that the PRMT5-specific inhibitor GSK3326595 is able to dramatically reduce ACE2 binding with RBD. Moreover, we discovered that meR671-ACE2 plays an important role in ACE2 binding with Spike1 of the SARS-CoV-2 Omicron, Delta, and Beta variants; and we found that GSK3326595 strongly attenuates ACE2 interaction with Spike1 of the SARS-CoV-2 Omicron, Delta, and Beta variants. Finally, SARS-CoV-2 pseudovirus infection assays uncovered that PRMT5-mediated meR671-ACE2 is essential for SARS-CoV-2 infection in human cells, and pseudovirus infection experiments confirmed that GSK3326595 can strongly suppress SARS-CoV-2 infection of host cells. Our findings suggest that as a clinical phase II drug for several kinds of cancers, GSK3326595 is a promising candidate to decrease SARS-CoV-2 infection by inhibiting ACE2 methylation and ACE2-Spike1 interaction.
1 INTRODUCTION
The COVID-19 epidemic has worsened from November 2019 to the present. COVID-19 disease is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).1, 2 According to the newest Worldometer statistical data released on August 12, 2022, there have been more than 593.43 million cases of COVID-19 worldwide, with over 6.449 million deaths due to the pandemic (https://www.worldometers.info/coronavirus/). SARS-CoV-2 infection is initiated by interaction of the viral spike protein with a receptor on the host cell surface.3 Various reports have confirmed that the Spike1 (S1) protein recognizes and binds with angiotensin-converting enzyme 2 (ACE2) to facilitate SARS-CoV-2 infection in humans.3-5 Crystal structure analysis has demonstrated that the receptor-binding domain (RBD) of S1 is responsible for interacting with ACE2.3, 4
Furthermore, an increasing number of SARS-CoV-2 mutant viruses have been reported, such as Beta, Delta, and Omicron variants.2, 6-13 In the past 10 months, the Omicron has become the major SARS-CoV-2 variant in the COVID-19 pandemic all over the world.9, 14-16 Many studies have shown that mutations allow the virus spread faster, leading to reduced vaccine protection.6, 10, 11, 17 It is particularly important to find several specific drugs to cure COVID-19, especially for critically ill patients. Many scientists believe that inhibition of RBD-ACE2 binding is a promising strategy to prevent SARS-CoV2 infection and/or cure COVID-19.3, 4 It has been reported that N-glycosylation may play a key role in RBD-ACE2 association.18-22 However, it has not yet been reported whether other posttranslational modifications (PTMs) (e.g., methylation, acetylation) of ACE2 also play an important role in RBD-ACE2 binding.
Proteins arginine methylation is a critical kind of PTMs of histone proteins and nonhistone proteins and is mediated by protein arginine methyltransferases (PRMTs).23 Our recent studies have shown that arginine methylation plays a key role in many biological functions, such as cancer metastasis,24-27 oxidative phosphorylation,28 oxidative stress resistance,29, 30 and cell senescence.31 PRMTs catalyse the formation of three kinds of arginine methylation: monomethyl-arginine (MMA), asymmetric dimethyl-arginines (ADMAs), and symmetric dimethyl-arginines (SDMAs). Protein argininemethyltransferase 5 (PRMT5), the predominant type II PRMT, mediates the formation of SDMAs in its substrates.32 The newest report reveals that monomethylation of PRMT7-mediated MAVS (mitochondrial antiviral-signaling protein) at the R52 site has a strong effect on the MAVS-mediated antiviral response.33 We sought to determine whether arginine methylation functions in SARS-CoV-2 infection of humans.
In this study, we found that PRMT5-mediated ACE2 symmetric dimethylation is necessary for SARS-CoV-2 infection in humans. We showed that PRMT5-mediated ACE2-R671 SDMA (meR671-ACE2) modification promotes binding of the SARS-CoV-2 RBD with ACE2 in vitro and in vivo. Then, we demonstrated that GSK3326595 (a PRMT5-specific inhibitor) dramatically inhibits the association of ACE2 with RBD. Moreover, we discovered that meR671-ACE2 plays an important role in the binding of SARS-CoV-2 Omicron, Delta, and Beta variant Spike1 with ACE2. We reveal that GSK3326595 (short for GSK595) is able to strongly decrease ACE2 interaction with Omicron, Delta, and Beta variant Spike1. Finally, we use SARS-CoV-2 pseudovirus infection experiments to show that PRMT5-mediated meR671-ACE2 is essential for SARS-CoV-2 infection in humans. In addition, we confirm that GSK3326595 strongly reduces SARS-CoV-2 infection in host cells based on SARS-CoV-2 pseudovirus infection assays. Our findings show that PRMT5 specific inhibitor GSK3326595 is a promising drug to diminish SARS-CoV-2 infection by inhibiting ACE2 methylation and ACE2-Spike1 interaction.
2 MATERIALS AND METHODS
2.1 Cells culture and transfection
HEK-293T, HK2, and BEAS-2B cells were cultured in Dulbecco's modified eagle medium (DMEM) medium with 10% fetal bovine serum (FBS). A549 cell was cultured in RPMI-1640 medium with 10% FBS. All the cell lines were obtained from the American Type Culture Collection or Cell library of the Chinese Academy of Sciences. HEK-293T and A549 cells were transfected with the indicated plasmids using Polyethyleneimine (PROTEINTECH) or Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer's instructions. The siNC control and siPRMT5 sequences were shown in the Supporting Information.
2.2 Plasmids and SARS-CoV-2 Spike1 proteins
Plasmid expressing Flag-ACE2-WT (Cat: HG10108-NF) and GFP-RBD (Cat: VG40592-ACGLN) were purchased from Sino Biological company. Flag-ACE2-R644K and Flag-ACE2-R671K mutant plasmids were constructed by Quick Mutation™ Plus Kit (Cat: D0208S) from Beyotime company. SARS-CoV-2 Spike S1-Omicron mutant (B.1.1.529 variant: A67V, H69del, V70del, T95I, G142D, V143del, Y144del, Y145del, N211del, L212I, ins214EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H) His Tag Protein (His-S1-Delta) (Cat: 40591-V08H41), SARS-CoV-2 Spike S1-Delta mutant (B.1.617.2 variant: T19R, G142D, E156G, 157-158 deletion, L452R, T478K, D614G, P681R) His Tag Protein (His-S1-Delta) (Cat: 40591-V08H23), and SARS-CoV-2 Spike S1-Beta mutant (B.1.351 variant: K417N, E484K, N501Y, D614G) His Tag Protein (His-S1-Beta) (Cat: 40591-V08H10) were purchased from Sino Biological company.
2.3 SARS-CoV-2 spike pseudovirus infection assays
SARS-CoV-2 (2019-nCoV) spike pseudovirus (Cat: PSV001) were purchased from Sino Biological company. The pseudovirus infection assays were conducted by the specification. In brief, 5 × 104 of HEK-293T-Flag-ACE2 or A549-Flag-ACE2 cells were seeded into 96-well plates and cultured at 37°C in an incubator with 5% CO2 for 12 h. First, the cells were pretreated with 100 μl of DMEM medium containing the corresponding concentration of the test substances (DMSO or GSK3326595) for 12 h. Second, 100 μl of DMEM medium containing 5 μl the SARS-CoV-2 spike pseudovirus was added into each well and incubated 6 h for further infection. Third, the DMEM medium containing the virus was removed and replaced with 200 μl of fresh DMEM medium, followed by continuous incubation at 37°C for 48 h. Fourth, the DMEM medium was subsequently aspirated from the 96-well plates. Finally, 20 μl of the cell lysate solution from the Luciferase assay system 100 assays (Promega; E1500) was added to each well, followed by 10 μl of the luminescence solution before the detection of luciferase luminescence.
2.4 Western blot, immunoprecipitation (IP), and coimmunoprecipitation (Co-IP) assays
Western blot, IP, and Co-IP were performed as described previously.34 Specific primary antibodies and beads related in this study are showed in the following: GAPDH (60004-1-AP; Proteintech), PRMT5 (18436-1-AP; Proteintech), GFP-Tag (KM8009; SUNGENE BIOTECH), Flag-Tag (KM8002; SUNGENE BIOTECH), ACE2 (92485S; CST), His-Tag (abs830002ss; Absin), Myc-Tag (GB12076; Servicebio), GST-Tag (KM8005; SUNGENE BIOTECH), Anti-GFP Nanobeads (KTSM1301; AlpaLife), Anti-Myc Nanobeads (KTSM1306; AlpaLife), Anti-Flag-beads (P2271-2ml; Beyotime).
2.5 GST-ACE2 purification, GST pull-down assay, and in vitro methylation assay
The DNA sequence for ACE2-WT (601−744 aa) and ACE2-R671K (601−744 aa) were amplified by polymerase chain reaction and subcloned into pGEX-4T-1 vector (GE Healthcare) for production of GST-ACE2 (601−744 aa)-WT/R671K (short for GST-ACE2-WT/R671K) fusion proteins. The GST-ACE2-WT/R671K proteins were expressed in bacteria (BL21) induced with IPTG (isopropyl-β-d-thio-galactoside). Then, we enriched and purified GST-ACE2-WT/R671K proteins by the Glutathione Agarose beads (Cat. No.: HY-K0211; MCE). The detailed protocols of in vitro methylation assay and GST pull-down assay were shown in Supporting Information.
2.6 Mass spectrometry (MS) analysis
We carried out IP Flag-ACE2 in Flag-ACE2 ectopic HEK-293T cells. Then, we got the Flag-ACE2 gel by protein SDS-PAGE electrophoresis. The Liquid chromatography-tandem MS analysis (LC-MS/MS, Q Exactive [Thermo Fisher Scientific]) and Easy-nLC 1000 [Thermo Fisher Scientific] machines were used) of ACE2 methylation modification were performed in the Applied Protein Technology, Shanghai, China.
2.7 Statistical analysis
The Student t-test was used to determine statistic significance of differences between groups. p < 0.05 was considered statistically significant. Data are presented as mean ± SD. Statistical analysis was performed using the GraphPad Prism software (GraphPad Software).
3 RESULTS
3.1 The PRMT5-specific inhibitor GSK3326595 attenuates ACE2-RBD interaction
To explore whether arginine methylation impacts SARS-CoV-2 infection in humans, we detected ACE2-RBD association in HEK-293T cells coexpressing ACE2 and RBD after treatment with AMI-1 (inhibition of ADMAs) or GSK3326595 (inhibition of SDMAs inhibitor, PRMT5-specific inhibitor). GSK3326595 (short for GSK595 in the following) dramatically decreased interaction between ACE2 and the RBD (Figure 1A). Given that GSK595 is a PRMT5-specific inhibitor and that PRMT5 catalyses protein SDMA modification, we speculated that GSK595 suppresses ACE2 binding with the RBD, probably by inhibiting PRMT5-mediated ACE2 SDMA modification. Using Co-IP assays, we found that ACE2 binds to PRMT5 in Flag-ACE2-overexpressing HEK-293T cells (Figure 1B). In addition, we also discovered that ACE2 has MMA and SDMA modifications (Figure 1B). Our data suggest that PRMT5 catalyses ACE2 SDMA modification.
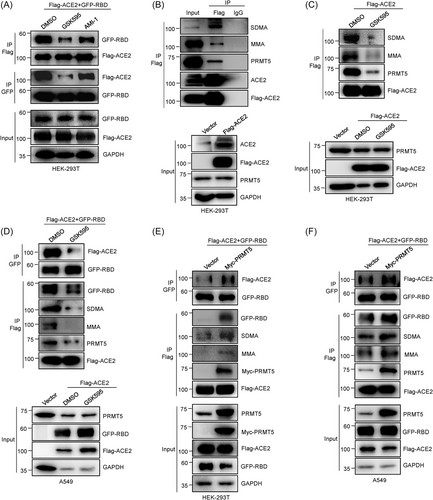
Subsequently, we used Co-IP Flag-ACE2 experiments in HEK-293T cells ectopically expressing Flag-ACE2 after treatment with PRMT5-specific inhibitor GSK595 and observed that GSK595 strongly decreases PRMT5 interaction with ACE2 (Figure 1C). Consistently, GSK595 significantly decreased ACE2 SDMA modification (Figure 1C). Moreover, we also confirmed that ACE2 SDMA modification, ACE2-RBD interaction, and ACE2-PRMT5 association were decreased after treated with GSK595 in Flag-ACE2 and GFP-RBD coexpression A549 cells (Figure 1D). Besides, ectopic expression of PRMT5 remarkably increased the ACE2-PRMT5 interaction and ACE2-SDMA modification in HEK-293T-(Flag-ACE2 + GFP-RBD) and A549-(Flag-ACE2 + GFP-RBD) cells, respectively (Figure 1E,F). Finally, our data revealed that overexpression of PRMT5 prominently strengthened the ACE2-RBD interaction (Figure 1E,F). These data indicate that PRMT5 catalyses ACE2 SDMA modification and that PRMT5-mediated ACE2-SDMA may play a key role in ACE2-RBD binding.
3.2 PRMT5-mediated ACE2-R671-SDMA is essential for ACE2-RBD interaction
To determine the exact arginine methylation sites of the ACE2 protein, we carried out MS methylation analysis of the Flag-ACE2 protein, which showed ACE2 sites R671 and R644 sites to be methylated (Figure 2A,B). To verify the major methylation site of ACE2, we constructed ACE2-R671K and ACE2-R644K mutant plasmids (mutant ACE2 cannot be methylated by PRMT5), overexpressed wild-type and mutant ACE2 plasmids in HEK-293T cells and detected methylation status changes. Our methylation assays revealed that only the ACE2-R671K mutant strongly inhibited the ACE2-PRMT5 association and decreased ACE2 SDMA modification in vivo (Figure 2C). Moreover, our in vitro methylation assays showed that PRMT5 directly methylated GST-ACE2 (Figure 2D). Our in vitro methylation data also confirmed that R671K mutant GST-ACE2 strongly decreased PRMT1-mediated ACE2 SDMA modification (Figure 2E). This data indicates that PRMT5 mainly catalyses SDMA modification of ACE2-R671 (meR671-ACE2).
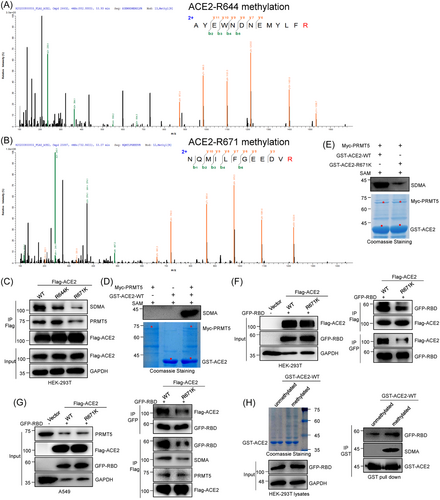
We then sought to explore the function of PTMT5-mediated meR671-ACE2 in ACE2-RBD binding. Our Co-IP assays revealed dramatically decreased ACE2-RBD interaction for the ACE2-R671K mutant compared with ACE2-WT in Flag-ACE2 and GFP-RBD coexpressing HEK-293T cells or A549 cells (Figure 2F,G). Besides, our GST pull-down assays also demonstrated that methylated-GST-ACE2-WT by PRMT5 increased ACE2-RBD interaction (Figure 2H). These data indicate that PRMT5-mediated ACE2-R671 SDMA modification has a great effect on the ACE2-RBD association.
3.3 Low concentration GSK3326595 can strongly inhibit ACE2-RBD interaction
Next, we explored the in-depth mechanism by which PRMT5-mediated meR671-ACE2 enhances ACE2-RBD interaction. Recently, several reports have shown that glycosylation of ACE2 plays a critical role in ACE2-RBD binding.18-21 Based on analysis of RBD-ACE2 binding domain crystal structure simulation, it was speculated that N-glycosylation of ACE2-N322 might promote SARS-CoV-2 infection by facilitating RBD-ACE2 association. Previous studies have reported that arginine methylation promotes crosstalk with other kinds of PTMs, such as phosphorylation and ubiquitination.24-26 Combined with our findings, we propose that PRMT5-mediated ACE2-R671 SDMA modification increases ACE2-RBD interaction by promoting ACE2-N322 N-glycosylation.
To test our hypothesis, we first constructed ACE2-N322Q mutant and ACE2-(R671K + N322Q) double-site mutant plasmids. Our Co-IP assays demonstrated decreased ACE2 binding with the RBD in both Flag-ACE2 single-site mutant (R671K or N322Q) and Flag-ACE2 double-site mutant (R671K + N322Q) groups compared with the Flag-ACE2-WT group (Figure 3A). Interestingly, we found that the amount of ACE2-RBD association decrease was almost the same in the three mutant groups (R671K group, N322Q group single-site ACE2 mutant groups, and [R671K + N322Q] double-site mutant group) compared with the ACE2-WT group (Figure 3A). It strongly means that ACE2-R671 methylation and ACE2-N322 glycosylation promote ACE2-RBD association dependent on the same pathway, which can explain why the single and the double mutant ACE2 decrease almost the same amount of ACE2-RBD binding.
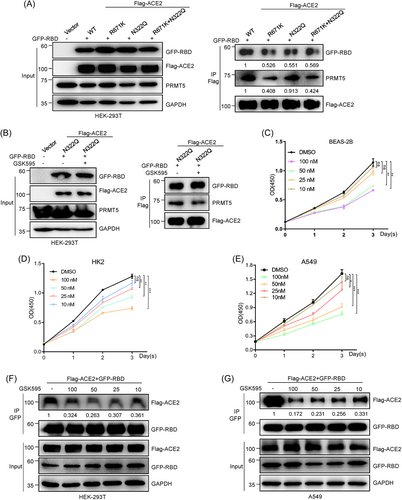
Moreover, our Co-IP experiments also showed that the level of ACE2-PRMT5 interaction did not change much between the ACE2-WT group and the N322Q mutant group (Figure 3A). This result suggests that ACE2-N322 N-glycosylation might have no effect on PRMT5-mediated meR671-ACE2 modification. In contrast, the level of ACE2 binding with the RBD was little changed after GSK3326595 treatment of Flag-ACE2-N322-overexpressing HEK-293T cells (Figure 3B). These data suggest that R671 methylation is the upstream and N322 glycosylation is the downstream in the R671 methylation and N322 glycosylation cascade facilitating ACE2-RBD interaction. Taken together, these data suggest that PRMT5-mediated meR671-ACE2 promotes RBD binding with ACE2, probably by facilitating ACE2-N322 N-glycosylation.
GSK3326595, a specific inhibitor of PRMT5, is a phase II clinical trial drug used for suppressing breast cancer cell proliferation,32, 35 and we wondered whether there is a concentration of GSK3326595 that can strongly repress ACE2 binding with the RBD but has little effect on cell proliferation. Some studies have demonstrated that lung tissues and the kidneys are mostly commonly infected by SARS-CoV-2.5, 12, 36 Hence, we first measured the proliferation ability of BEAS-2B (human bronchial epithelial cell), HK2 (human renal tubular epithelial cell), and A549 (human lung cancer cell) cells after treatment with GSK3326595 by CCK-8 assays (Figure 3C−E). Although GSK3326595 at 25 nM had a very minor effect on cell proliferation, ACE2 interaction with the RBD was remarkably decreased after treated with GSK3326595 at 25 nM concentration in Flag-ACE2 and GFP-RBD coexpressing HEK-293T cells and A549 cells (Figure 3F,G). This result indicates that 25 nM GSK3326595 may be the ideal concentration for inhibiting SARS-CoV-2 infection in humans.
3.4 GSK3326595 inhibits SARS-CoV-2 Omicron and other variants Spike1 binding with ACE2
We showed that PRMT5-mediated ACE2-R671 SDMA modification is essential for RBD binding with ACE2 and that GSK3326595 is able to prevent ACE2-RBD interaction. Recent studies have found that several SARS-CoV-2 variants, such as the Omicron, Delta, and Beta variants, exhibit much higher viral transmissibility and a much higher level of escape from SARS-CoV-2 vaccine-induced immunity.2, 6, 9, 11, 17 For instance, the SARS-CoV-2 Delta variant has spread worldwide since it was first identified in India in October 2020.8, 17 The recently report shows that the SARS-CoV-2 Omicron variant is spreading rapidly from South Africa to the worldwide in the past 10 months since it was found in November 2021.9, 14-16 Therefore, we wondered whether PRMT5-mediated ACE2-R671 methylation also plays a critical role in ACE2 binding with Spike1 of SARS-CoV-2 Omicron and other variants.
We first detected the ACE2 and S1 interaction by Co-IP assays after incubating Flag-ACE2-WT/R671K HEK-293T cell lysates with His-S1-Omicron or His-S1-Delta or His-S1-Beta protein. ACE2-R671K group strongly inhibited S1 binding with ACE2 for the Omicron, Delta, and Beta variants compared with ACE2-WT (Figure 4A−C). Then, we found that GSK3326595 can strongly prevent the S1 proteins of the above mentioned three SARS-CoV-2 variants (Omicron, Delta, and Beta) binding with Flag-ACE2-WT, as based on Co-IP experiments (Figure 4D−F). Moreover, we showed that the reduced levels of His-S1-Omicron, His-S1-Delta, and His-S1-Beta binding with Flag-ACE2-WT correlated positively with the dose of GSK3326595 concentration (Figure 4G−I). Our data reveal that GSK3326595 is able to strongly prevent the above mentioned three SARS-CoV-2 variants S1 proteins from interacting with Flag-ACE2-WT, especially for His-S1-Omicron and His-S1-Delta variants even at 25 μM concentration (Figure 4G,H). These data demonstrate that PRMT5-mediated meR671-ACE2 is required for the SARS-CoV-2 Omicron, Delta, and Beta variants to interact with ACE2 and that GSK3326595 inhibits their interaction with ACE2. This strongly suggests that GSK3326595 may protect humans from SARS-CoV-2 Omicron, Delta, and Beta variant infection.
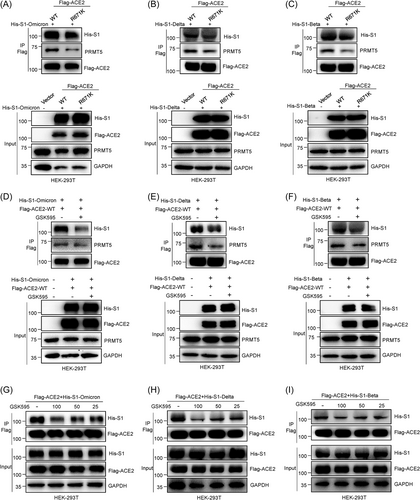
3.5 GSK3326595 dramatically inhibits SARS-CoV-2 spike pseudovirus infection host cells
To assess the direct function of ACE2-R671 SDMA in SARS-CoV-2 infection of human host cells, we carried out SARS-CoV-2 spike pseudovirus infection in ACE2-overexpressing HEK-293T cells. Strikingly, we found that the SARS-CoV-2 spike pseudovirus infection rate decreased with increasing GSK3326595 concentration (Figure 5A). Our data also revealed that at the concentration of 25 nM, GSK3326595 can prevent almost 70% of pseudovirus infection of host cells (Figure 5A). And our SARS-CoV-2 spike pseudovirus infection assays got the similar results in ACE2-overexpressing A549 cells after treated GSK3326595 with different concentrations (Figure 5B). Furthermore, we detected the SARS-CoV-2 spike pseudovirus infection after knockdown PRMT5 in ACE2-overexpressing HEK-293T cells and ACE2-overexpressing A549 cells. Our results discovered that silencing of PRMT5 expression decreased pseudovirus infection of HEK-293T cells and A549 cells (Figure 5C−F). These data strongly indicate that PRMT5 might play a critical part in SARS-CoV-2 infection into host cells.
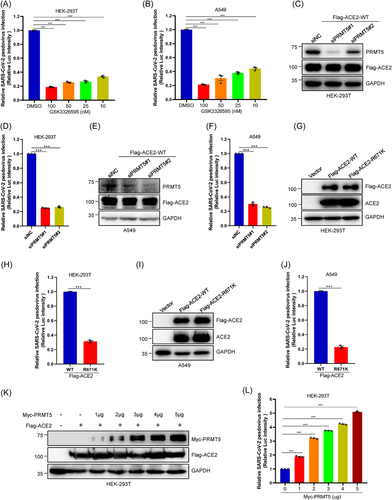
Subsequently, we also confirmed that the Flag-ACE2-R671K mutant significantly suppresses SARS-CoV-2 spike pseudovirus infection of host cells both in HEK-293T cells and A549 cells (Figure 5G−J). Consistently, the SARS-CoV-2 spike pseudovirus infection rate elevated with increasing PRMT5 protein expression (Figure 5K,L). Taken together, these data clearly indicate that PRMT5-mediated ACE2-R671 SDMA modification is necessary for SARS-CoV-2 infection. Our results also strongly indicate that GSK3326595 may have a great effect on preventing SARS-CoV-2 infection in humans.
4 DISCUSSION
In this study, we demonstrate that PRMT5-mediated ACE2-R671 SDMA is necessary for ACE2 binding with the RBD of the SARS-CoV-2 Spike1 protein (Figures 1, 2). In terms of the mechanism, our data suggest that PRMT5-mediated meR671-ACE2 facilitates ACE2-RBD interaction probably by promoting ACE2-N322 N-glycosylation (Figure 3A,B). Moreover, we discovered that GSK3326595 (a PRMT5-specific inhibitor) is able to prevent the Spike1 protein of the SARS-CoV-2 Omicron, Delta, and Beta variants from interacting with ACE2 (Figure 4). Finally, we used pseudovirus infection assays to confirm that GSK3326595 strongly inhibits SARS-CoV-2 infection of host cells (Figure 5).
A recent report showed that PRMT7-mediated MAVS-R52 monomethylation can suppress antiviral innate immunity in humans, revealing that MAVS-R52 methylation inhibits MAVS interaction with TRIM31 and RIG-I, which represses MAVS aggregation and activation.33 Upon virus infection, MAVS promotes PRMT7 ubiquitin-proteasome degradation by recruiting the E3 ligase SMURF1 to bind PRMT7, which increases MAVS activation and subsequent antiviral immunity. This study revealed that arginine methylation regulates human antiviral defense. Indeed, we found that PRMT5 catalyses ACE2-R671 dimethylation and plays a key role in SARS-CoV-2 virus infection of host cells by regulating ACE2-RBD interaction. Thus, arginine methylation may have a great effect on controlling virus infection, and regulation of PRMT protein enzyme activity may be a way to strengthen antiviral capacity.
The SARS-CoV-2 Omicron and Delta variant is spreading fast in many countries and regions.1, 2, 9, 15 The newest WHO statistical data have confirmed that Omicron variant is becoming the major SARS-CoV-2 form spreading in the world in the past several months.9 The Delta variant has a much higher ability of immune escape in the presence of many kinds of SARS-CoV-2 vaccines.6, 11 The Omicron variant has a much higher spread ability.14, 16 Therefore, it is very important to find drugs to fight SARS-CoV-2 variants, especially the Omicron and Delta variants. Our research shows that GSK3326595 at a very low concentration is able to successfully attenuate ACE2 binding with Spike1 of the SARS-CoV-2 Omicron, Delta, and Beta variants. As an oral effective specific inhibitor of PRMT5, GSK3326595 is a clinical phase II drug for myelodysplastic syndromes and patients with acute myeloid leukemia and breast cancer.35 Based on our findings in this study, we believe that GSK3326595 is a promising specific drug that will inhibit SARS-CoV-2 infection in humans and cure COVID-19 patients. However, more evidence is needed. In the future, we will attempt to carry out an animal study in hACE2 transgenic mice and a clinical trial using the GSK3326595 drug in COVID-19 patients infected with the SARS-CoV-2 Omicron and Delta variants.
In summary, our study reveals that PRMT5-mediated ACE2 dimethylation at the R671 site is essential for SARS-CoV-2 Spike1 binding with ACE2 and subsequent infection of host cells (Figure 6). Moreover, we show that the PRMT5-specific inhibitor GSK3326595 is a promising drug to reduce SARS-CoV-2 infection.

AUTHOR CONTRIBUTIONS
Study conception and experimental design, collection and analysis of data, and manuscript writing: Zhongwei Li, Hongmei Yong, and Wenwen Wang. Collection and analysis of data: Zhongwei Li, Yue Gao, Wenwen Wang, Xintian Chen, and Pengfei Wang. Study conception, design, and supervision: Zhongwei Li, Junnian Zheng, and Jin Bai. Supervision, manuscript writing, and final approval of the manuscript: Zhongwei Li, Junnian Zheng, and Jin Bai.
ACKNOWLEDGMENTS
We thank Dr. Yibo Wang and Dr. Hongyuan Li (Laboratory of Chemical Biology, Changchun Institute of Applied Chemistry, Chinese Academy of Sciences, Changchun, Jilin, China) for their helpful suggestions. This study was supported by grants from the National Natural Science Foundation of China (82173060); the Excellent Youth Foundation of Jiangsu Province, China (BK20220119); the Major Project of University Natural Science Foundation of Jiangsu Province, China (22KJA310006); the National Postdoctoral Research Funds of China (2019M651971 and 2021T140577); and the Qinglan Project of Jiangsu.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




