Rapid and reliable inactivation protocols for the diagnostics of emerging viruses: The example of SARS-CoV-2 and monkeypox virus
Abstract
The emergence and sustained transmission of novel pathogens are exerting an increasing demand on the diagnostics sector worldwide, as seen with the ongoing severe acute respiratory coronavirus 2 (SARS-CoV-2) pandemic and the more recent public health concern of monkeypox virus (MPXV) since May 2022. Appropriate and reliable viral inactivation measures are needed to ensure the safety of personnel handling these infectious samples. In the present study, seven commercialized diagnosis buffers, heat (56°C and 60°C), and sodium dodecyl sulfate detergent (2.0%, 1.0%, and 0.5% final concentrations) were tested against infectious SARS-CoV-2 and MPXV culture isolates on Vero cell culture. Cytopathic effects were observed up to 7 days postinoculation and viral load evolution was measured by semiquantitative polymerase chain reaction. The World Health Organization recommends an infectious titer reduction of at least 4 log10. As such, the data show efficacious SARS-CoV-2 inactivation by all investigated methods, with >6.0 log10 reduction. MPXV inactivation was also validated with all investigated methods with 6.9 log10 reductions, although some commercial buffers required a longer incubation period to yield complete inactivation. These results are valuable for facilities, notably those without biosafety level-3 capabilities, that need to implement rapid and reliable protocols common against both SARS-CoV-2 and MPXV.
1 INTRODUCTION
The zoonotic emergence and sustained human-to-human transmission of novel pathogens are of great concern for the implementation of safety measures when handling potentially infectious samples. The uncertain dissemination of these novel pathogens requires the capacity of health facilities to respond rapidly and at scale with adaptable security measures.
This was made apparent with the ongoing severe acute respiratory coronavirus 2 (SARS-CoV-2) pandemic from late 2019 and proves to be of more recent interest with the monkeypox virus (MPXV) since May 2022. Indeed, the latter is the cause of the largest multicountry outbreak outside of endemic areas of western and central Africa with over 47 600 confirmed or probable cases in 98 countries as of August 29, 2022.1, 2 SARS-CoV-2 and MPXV differ both in terms of viral structure and of global health alert. SARS-CoV-2 is a single-stranded RNA virus responsible for the COVID-19 pandemic with an unprecedented scale of human infection. MPXV is a double-stranded DNA virus for which the disease is known but the current epidemiology diverges from trends in the past. Despite these differences, in both cases, a viral pathogen is spreading at relatively high rates and is coming into contact with a greater reach of a global population. As such, medical and nonmedical structures receiving potentially infectious SARS-CoV-2 and MPXV samples and all actors implicated in the diagnostic and downstream processes need to be made aware of effective inactivation methods.
For these reasons, the objective of this study is to evaluate the inactivation efficacy of simple chemical and physical methods to secure the safety of handling infectious SARS-CoV-2 and MPXV specimens.
2 MATERIAL AND METHODS
2.1 Viruses
A SARS-CoV-2 (family Coronaviridae genus Betacoronavirus species SARS-related coronavirus) isolate belonging to the 20C clade (GISAID, accession number EPI_ISL_640002) was obtained by the French National Reference Centre for Respiratory Viruses. A MPXV (family Poxviridae subfamily Chordopoxvirinae genus Orthopoxvirus species monkeypox virus) isolate belonging to clade II (NCBI, accession number ON622722.2) was obtained by the virology lab of the Hospices Civils de Lyon. Both viruses were propagated on Vero cells (ATCC©, CCL-81) with Eagle's minimum essential medium culture media, supplemented with 2% penicillin–streptomycin + 1% l-glutamine + 2% fetal bovine serum.
All virus propagation experiments were performed in the biosafety level-3 (BSL-3) laboratory in compliance with the Microorganism and Toxin legislation at the Institut des Agents Infectieux—Hospices Civils de Lyon.
2.2 Inactivation conditions
The buffers from commercialized diagnostic kits tested on both SARS-CoV-2 and MPXV are the BD Max buffer (Becton, Dickinson), Cobas® lysis buffer (Roche, DE), Cobas® viral transport medium (Roche), NucliSENS® EMAG® lysis buffer (bioMérieux), Maxwell lysis buffer (Promega), Panther Fusion™ lysis buffer (Hologic®), and Sun-Trine® viral transport medium (SunTrine® Biotechnologies). Heat inactivation was carried out with a dry-heat oven. SARS-CoV-2 and MPXV were tested at 56°C for 30 and 10 min, and at 60°C for 1 h, 30 min, and 10 min. Sodium dodecyl sulfate (SDS; CAS n°151-21-3) was used at final concentrations of 2.0%, 1.0%, and 0.5% for 30 and 10 min.
2.3 Inactivation procedure
For commercial buffers, 100 µl of virus culture sample were added to 900 µl commercial buffer and incubated at room temperature for 10 min. An additional condition of 30 min of incubation was tested for commercial buffers demonstrating a less efficacious inactivation at 10 min. For the heat protocols, 100 µl of culture sample were added to 900 µl of supplemented culture media and incubated at appropriate temperatures and times. For the SDS protocols, 100 µl of culture sample were added to 900 µl of supplemented culture media with the appropriate quantity of SDS for the final concentrations described above and then incubated at room temperature for appropriate times.
Once the contact period was completed, each inactivation condition was diluted through 10-fold serial dilutions in supplemented culture media and then inoculated in triplicate on confluent Vero cells seeded in 96-well plates. To account for the eventual cytotoxic property of the chemical agents, control wells were prepared for all inactivation conditions in the absence of the virus. The plates were incubated at 36°C under 5% CO2 for 96 h, with an extended period of incubation of 7 days for MPXV. Cytotoxicity and virus growth was monitored by optical microscopy. To confirm the observation of cytopathic effects (CPEs), the supernatant of each condition was sampled after the contact period (D0) and then again after 96 h of incubation (DX) for RNA extraction and quantitative reverse transcription-polymerase chain reaction (qRT-PCR) detection with the TaqPath COVID-19 CE-IVD Kit (ORF1ab target gene; Thermo Fisher Scientific) or after 7 days (DX) for DNA extraction and qPCR detection with the TaqMan Monkeypox Detection Assay (Thermo Fisher Scientific) on a QuantStudio 5 System (Applied Biosystems).3 Virus culture growth was considered positive when, after at least 96 h of incubation, characteristic CPEs were observed and a viral load increase by at least 1.0 log10 was detected; for analysis, polymerase chain reaction (PCR) detection limit cutoff was set at 40 PCR cycle thresholds (Ct). [∆log10 = average Ct at given dilution at D0 − Ct of tested condition at DX)/3.33, as 1log10 ≈ 3.3 Ct].
Infectious titers were calculated with the Spearman–Karber method.3 The analysis of virus inactivation was based on recommendations of the European norm NF EN 14476-A2,4 where the difference between infectious titers of untreated and treated conditions constitutes the log10 reduction value.
3 RESULTS
3.1 SARS-CoV-2
3.1.1 Untreated virus control
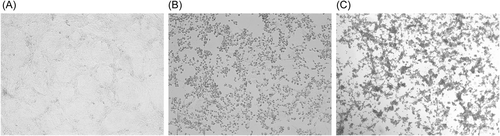
The untreated SARS-CoV-2 control yielded typical CPE, defined by morphological changes such as rounding of cells and lysis of the cell monolayer (Figure 1). This SARS-CoV-2 cell culture passage resulted in an average infectious titer of 6.0 log10 median tissue culture infectious dose (TCID50/ml; Table 1). Semiquantitative qRT-PCR data supported these findings, with active virus genome replication detected for all CPE-positive wells (Supporting Information: Data 1).
| Inactivation method | Log10 TCID50/ml | LRV |
|---|---|---|
| Virus control (untreated) | 6.0 | NA |
| Commercial buffers (10 min) | ||
| BD Max buffer | 0.0 | >6.0 |
| Cobas® lysis buffer | 0.0 | >6.0 |
| Cobas® viral transport medium | 0.0 | >6.0 |
| Maxwell lysis buffer | 0.0 | >6.0 |
| NucliSENS® EMAG® lysis buffer | 0.0 | >6.0 |
| Panther Fusion™ lysis buffer | 0.0 | >6.0 |
| SunTrine® viral transport medium | 0.0 | >6.0 |
| Heat (1 h) | ||
| 60°C | 0.0 | >6.0 |
| Heat (30 min) | ||
| 60°C | 0.0 | >6.0 |
| 56°C | 0.0 | >6.0 |
| Heat (10 min) | ||
| 60°C | 0.0 | >6.0 |
| 56°C | 0.0 | >6.0 |
| SDS detergent (30 min) | ||
| 2.0% | 0.0 | >6.0 |
| 1.0% | 0.0 | >6.0 |
| 0.5% | 0.0 | >6.0 |
| SDS detergent (10 min) | ||
| 2.0% | 0.0 | >6.0 |
| 1.0% | 0.0 | >6.0 |
| 0.5% | 0.0 | >6.0 |
- Abbreviations: LRV, log10 reduction value; NA, not applicable; SARS-CoV-2, severe acute respiratory coronavirus 2; SDS, sodium dodecyl sulfate; TCID50/ml, median tissue culture infectious dose.
3.1.2 Commercial buffers
Cytotoxicity was observed for all commercial buffers tested at more concentrated dilutions. Nevertheless, 10-fold serial dilutions provided a cytotoxic limit from which cytotoxicity was no longer observed. No CPE was observed for any commercial buffers (Table 1). Semiquantitative qRT-PCR data supported the virus culture results, with no active genome replication detected for CPE-negative wells (Supporting Information: Data 1). All commercial buffers tested herein, therefore, yield a >6 log10 reduction of active SARS-CoV-2 replication.
3.1.3 Heat
No CPE was observed for conditions treated with the heat protocols (Table 1). Semiquantitative qRT-PCR data supported these findings (Supporting Information: Data 1). All heat protocols therefore also yield a >6 log10 reduction of active SARS-CoV-2 replication.
3.1.4 SDS detergent
Cytotoxicity was observed for all initial SDS concentrations, but 10-fold serial dilutions provided a cytotoxic limit from which cytotoxicity was no longer observed. No CPE was observed for any SDS concentration (Table 1). As with the previous inactivation methods, qRT-PCR data supported these findings, with no active genome replication detected for CPE-negative wells (Supporting Information: Data 1). All SDS concentrations tested, therefore, yield a >6 log10 reduction of active virus replication.
3.2 Monkeypox virus
3.2.1 Untreated virus control
The untreated MPXV control yielded typical CPE on Vero cells (Figure 1). This MPXV cell culture passage resulted in an average infectious titer of 6.9 log10 TCID50/ml (Table 2). The cell culture results were confirmed with semiquantitative qPCR data, with active virus genome replication detected for all CPE-positive wells (Supporting Information: Data 2).
| Inactivation method | Log10 TCID50/ml | LRV |
|---|---|---|
| Virus control (untreated) | 6.9 | NA |
| Commercial buffers (10 min) | ||
| BD Max buffer | 0.0 | >6.9 |
| Cobas® lysis buffer | 4.5 | 2.4 |
| Cobas® viral transport medium | 2.5 | 4.4 |
| Maxwell lysis buffer | 0.0 | >6.9 |
| NucliSENS® EMAG® lysis buffer | 4.2 | 2.7 |
| Panther Fusion™ lysis buffer | 3.9 | 3.0 |
| SunTrine® viral transport medium | 0.0 | >6.9 |
| Commercial buffers (30 min) | ||
| Cobas® lysis buffer | 0.0 | >6.9 |
| Cobas® viral transport medium | 0.0 | >6.9 |
| NucliSENS® EMAG® lysis buffer | 0.0 | >6.9 |
| Panther Fusion™ lysis buffer | 0.0 | >6.9 |
| Heat (1 h) | ||
| 60°C | 0.0 | >6.9 |
| Heat (30 min) | ||
| 60°C | 0.0 | >6.9 |
| 56°C | 0.0 | >6.9 |
| Heat (10 min) | ||
| 60°C | 0.0 | >6.9 |
| 56°C | 0.0 | >6.9 |
| SDS detergent (30 min) | ||
| 2.0% | 0.0 | >6.9 |
| 1.0% | 0.0 | >6.9 |
| 0.5% | 0.0 | >6.9 |
| SDS detergent (10 min) | ||
| 2.0% | 0.0 | >6.9 |
| 1.0% | 0.0 | >6.9 |
| 0.5% | 0.0 | >6.9 |
- Abbreviations: LRV, log10 reduction value; MPXV, monkeypox virus; NA, not applicable; SDS, sodium dodecyl sulfate; TCID50/ml, median tissue culture infectious dose.
3.2.2 Commercial buffers
Cytotoxicity was observed for all commercial buffers tested at more concentrated dilutions. Nevertheless, serial dilutions provided a cytotoxic limit from which cytotoxicity was no longer observed. CPE and PCR data showed a less efficacious inactivation for four commercial buffers (Cobas® lysis buffer, Cobas® viral transport medium, NucliSENS® EMAG® lysis buffer, and Panther Fusion™ lysis buffer), yielding titers greater than 2 log10 TCID50/ml. A complete absence of CPE was observed for all other buffers (Table 2). Semiquantitative qPCR data supported these findings. No active genome replication was detected for CPE-negative wells, and PCR data allowed the clarification of a number of ambiguous cells culture wells, where cytotoxicity and CPE were difficult to validate (Supporting Information: Data 2). An additional protocol was included for Cobas® lysis buffer, Cobas® viral transport medium, NucliSENS® EMAG® lysis buffer, and Panther Fusion™ lysis buffer with a 30-min incubation period. This protocol produced similar cytotoxicity results as with 10 min and yielded complete inactivation with 0 log10 TCID50/ml titers. Taken together, the data show that all commercial buffers tested herein achieve a >6.9 log10 reduction inactivation against MPXV; 10 min of incubation is adequate for BD Max buffer, Maxwell lysis buffer, and SunTrine® viral transport medium, while 30 min is necessary for Cobas® lysis buffer, Cobas® viral transport medium, NucliSENS® EMAG® lysis buffer, and Panther Fusion™ lysis buffer.
3.2.3 Heat
No CPE was observed for conditions treated with heat protocols (Table 1). Semiquantitative qPCR data supported these findings (Supporting Information: Data 2). All heat protocols also yield a >6.9 log10 reduction of active MPXV replication.
3.2.4 SDS detergent
Cytotoxicity was observed for all initial SDS concentrations, but the serial dilutions provided a cytotoxic limit from which cytotoxicity was no longer observed. No CPE was observed for any SDS concentration (Table 1). As with the previous inactivation methods, qPCR data supported these findings, with no active genome replication detected for CPE-negative wells (Supporting Information: Data 2). All SDS concentrations, therefore, yield a >6.9 log10 reduction of active virus replication.
4 DISCUSSION
According to the European NF EN 14476-A2 and the World Health Organization (WHO) recommendations on robust and reliable viral safety, the standard acceptance criterion for virucidal substances is their ability to remove or inactivate 4 log10 or more amounts of virus.4, 5 Hence, our data explicitly demonstrate inactivation efficacy against both SARS-CoV-2 and MPXV for a number of investigated methods.
This information is particularly valuable for facilities, notably those without BSL-3 capabilities, that would need to implement rapid and reliable protocols during an evolving public health situation, all while reducing contamination risk. Indeed, WHO, regional, and local recommendations for clinical management of potential cases have been published for both SARS-CoV-2 and MPXV.6, 7 Our work takes these precautions further with direct inactivation of the pathogen.
Inactivation efficacy has been previously described on heat inactivation against influenza8 and SARS-CoV 20039 viruses as well as on detergent inactivation against enveloped herpes simplex and human immunodeficiency virus.10 In the present study, we find similar findings for heat and detergent inactivation against SARS-CoV-211, 12 but with more SDS concentrations tested and the addition of commercial buffers widely used in Europe during the COVID-19 pandemic. To date, there is little knowledge of inactivation specifically against monkeypox. In fact, most relevant documentation stems from experiments on the vaccinia virus, belonging to the same Orthopoxvirus genus as MPXV. Previous work has demonstrated the efficiency of heat, commercial buffers, or household detergents against the vaccinia virus, notably for surface decontamination.13, 14 To our knowledge, our work is one of the first in testing inactivation protocols specifically against MPXV culture isolates with widely available material and easy setup. Batéjat et al. also recently reported heat inactivation against MPXV, supporting our findings, and even demonstrating inactivation at 70°C for less than 5 min.15
As different forms of specimens, including sputum, plasma, and stool can be handled, the methods to inactivate these different samples can vary greatly, but the most common are heat and detergents.11, 14, 16 Numerous studies have even described SARS-CoV-2 inactivation efficacy with less traditional protocols including ophthalmic solutions or repurposed therapeutic agents.17, 18 Nevertheless, while these innovative efforts contribute to our growing knowledge of SARS-CoV-2, not all protocols may be readily available options to many medical or nonmedical institutions and facilities, and they have not been tested in the context of other emerging pathogens, such as MPXV. Heat and SDS seem to be easily accessible and simple procedures for inactivation, with our data showing efficacy by SDS alone at concentrations as low as 0.5% when others reported higher concentrations and/or in combination with other agents.12, 19
All described methods may also impact the structural integrity of the virus genome and protein structures and therefore constitutes another factor to consider when adopting an appropriate inactivation protocol. For example, SARS-CoV-2 genome instability with RT-PCR detection has been reported, with high heat inactivation showing an inappropriate protocol at 92°C.20 This protocol may have a different impact on MPXV, as DNA is more stable and less sensitive to degradation.
5 CONCLUSION
Overall, our study demonstrates the effective virus inactivation of SARS-CoV-2 and MPXV replication by various commercial buffers, heat, and SDS detergent. To conduct SARS-CoV-2 and MPXV research at lower biocontainment levels, a selection of commercial buffers, 56°C or 60°C for 10 min, and SDS as low as 0.5% could be effective methods of inactivation, but the choice of the protocol should always be in adequation with the downstream biological process and is still contingent on respecting maximum risk management.
AUTHOR CONTRIBUTIONS
Alexandre Gaymard is responsible for the conception and design of the study. Martine Valette, Maude Bouscambert-Duchamp, and Emilie Frobert provided access to the monkeypox virus isolate and biosafety level-3 laboratory. Martine Valette, Vanessa Escuret, and Bruno Lina provided access to the severe acute respiratory coronavirus 2 isolates. Geneviève Billaud provided commercial buffers and access to the molecular biology platform. Grégory Quéromès, Alexandre Gaymard, and Antoine Oblette contributed to sample preparation and data collection. Grégory Quéromès and Alexandre Gaymard were involved in the data analysis and drafting of the original manuscript. All authors revised the manuscript and approved the final version.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
1 DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author, upon reasonable request.




