Longitudinal follow-up of HPV16 sequence after cervical infection: Low intrahost variation and no correlation with clinical evolution
Abstract
Human papillomavirus (HPV) 16 exhibits different variants that may differ in their carcinogenic risk. To identify some high-risk variants, we sequenced and compared HPV16 whole genomes obtained from a longitudinal cohort of 34 HPV16-infected women who had either spontaneously cleared their infection (clearance group or “C”), or developed cervical high-grade lesions following a viral persistence (group persistence or “P”). Phylogenetic analysis of paired samples obtained at the beginning (C0 or P0) and at the end (C2 or P2) of the follow-up (median intervals between C0–C2 and between P0–P2 were 16 and 36.5 months, respectively) revealed a low genetic variability within the host compared to the genetic interhost diversity. By comparing our HPV16 sequences to a reference sequence, we observed 301 different substitutions, more often transitions (60.9%) than transversions (39.1%), that occurred throughout the viral genome, but with a low frequency in E6 and E7 oncogenes (10 and 9 substitutions), suggesting a high conservation of these genes. Deletions and insertions were mostly observed in intergenic regions of the virus. The only significant substitution found between the subgroups C2 and P2 was observed in the L2 gene (L330F), with an unclear biological relevance. Our results suggest a low longitudinal intrahost evolution of HPV16 sequences and no correlation between genetic variations and clinical evolution.
1 INTRODUCTION
Human papillomaviruses (HPVs) are sexually transmitted viruses, infecting epithelial cells of the lower anogenital tract of both males and females as well as of the oropharynx, oral cavity, and larynx.1, 2 Among the estimated 400 different HPV genotypes,3 13 are considered high-risk (hr) types due to their major involvement in promoting cancers at anogenital and oropharyngeal sites.4 HPV16 is by far the most potent carcinogenic genotype, detected in nearly two-thirds of cervical cancers and in a large part of other HPV-induced cancers.1, 5 Within the HPV16 genotype, different genetic variants, sharing more than 98% identity, may be identified. These HPV16 variants can be classified into four lineages (A, B, C, and D), which are suspected to vary in their carcinogenic risk.6, 7 A more accurate determination of the carcinogenic risk associated with each variant (or to each specific genetic variation) could help adapt the medical care in case of HVP16 detection in a cervical sample. The recent emergence of Next Generation Sequencing (NGS) has offered new perspectives for more accurate identification and characterization of variants, harboring specific genetic patterns.6, 8
Here, to identify specific HPV16 variants associated with the development of cervical (pre-)cancers, we sequenced HPV16 whole genomes obtained from two groups of HPV16 positive women with longitudinal follow-up in our medical institution, who had either spontaneously cleared the virus or who were persistently infected with a progression to the high-grade cervical lesion (HSIL). We describe the genetic variations observed in each group and aim to associate these variations with the clinical outcome.
2 MATERIALS AND METHODS
2.1 Study population
Adult women (≥18 years) with a history of cervical samples performed for longitudinal cervical cancer screening in the Department of Gynaecology and Obstetrics, Besançon University Hospital (France), were eligible. All samples were characterized for routine conventional cytology and HPV testing (see below). Inclusion criteria of patients were as follows: having at least one sample diagnosed as HPV16+ with a normal cervical smear or with a smear showing a mild cytological dysplasia (atypical squamous cells of undetermined significance, or low-grade squamous intraepithelial lesion [LSIL]) at the beginning of the follow-up, availability of all clinical and biological data (i.e., age, contraception, parity, tobacco, virological and cytological characterization of cervical samples, histological characterization of cervical lesions, treatment modalities), availability of all collected cervical samples stored into a biobank for which a declaration of preparation and storage of human samples for research use has been sent to the Ministère de l'Enseignement Supérieur et de la Recherche (n° DC-2014-2086). Patient inclusion, data collection, and NGS analysis were performed retrospectively.
Patients were subsequently classified into two groups (Supporting Information: Figure S1): (i) the first group of patients, named “clearance” or “C,” who had spontaneously cleared their HPV16 infection without any cervical lesion. In this group, the first (facultative) and last cervical samples that harbored a detectable HPV16 were called C0 and C2, respectively. (ii) The second group of patients, named “persistence” or “P,” for whom the detection of HPV16 had persisted during the follow-up, leading to the development of HSIL according to the Lower Anogenital Squamous Terminology classification.9 All the high-grade lesions were histologically confirmed. In this group, the first and last HPV16+ cervical samples taken into account were called P0 and P2, respectively, with P2 obtained at the time of detectable precancerous lesions.
2.2 Clinical samples
At each visit, two samples of exfoliated cells were obtained from the cervix: a first sample was collected with a Cytobrush® Plus (Medscand Medical) for conventional cytology and a second sample for HPV testing was collected with the Digene Cervical Sampler (Qiagen) immediately placed in 1 ml of a specific transport medium (Digene Specimen Transport Medium; Qiagen), transported to the laboratory within 24 h, and stored at 4°C until further analysis by the Digene Hybrid Capture 2 High-Risk HPV DNA Test (HC2) (Qiagen) and by an in-house HPV16 and HPV18 quantitative real-time polymerase chain reaction (PCR), as previously described.10 Samples containing denatured DNA used for HC2 testing were subsequently stored at −20°C until NGS analysis.
2.3 NGS protocol and data analysis
2.3.1 Library preparation and sequencing
Libraries were prepared from the frozen stored samples containing the denatured DNA using the Ampliseq library Plus for Illumina Kit (Illumina), according to the manufacturer's instructions. An initial step of neutralization with potassium acetate (3 M) and acetic acid (5 M) was necessary due to the presence of a basic solution added during the HC2 testing. DNA was extracted using the QIAmp DNA Mini Kit (Qiagen). Then, 20 ng of DNA were amplified by multiplex PCR using 26 paired primers covering the whole genome of HPV16.11 Thermocycling conditions were 99°C for 2 min, followed by 20 cycles of 99°C for 15 s and 60°C for 4 min. Amplicons were then digested using FuPa Reagent (ThermoFisher), indexes were ligated, and a new amplification step was performed. Sequencing was performed on MiSeq (Illumina Inc.) using the MiSeq Reagent Kit, according to the manufacturer's instructions.
2.3.2 Pipeline and data analysis
Amplicon sequencing data were analyzed using a metabarcoding approach. For this purpose, contigs were generated with Mothur (v1.43.0) and the Needleman alignment method. To remove chimera and artifact sequences, unique contigs were aligned against the HPV16 reference genome (K02718.1) with BLAT (v36x5) using 40 as a max-intron parameter. To further identify the most representative sequences in each sample, Swarm (v2.2.2) was used to cluster the contigs, and only sequences seen at least two times were kept. Those sequences were then compared to the HPV16 reference genome (K02718.1) to identify single nucleotide polymorphisms (SNPs) and indels in each sample with a homemade pipeline written with Python (3.7.6), matplotlib (3.1.1), pandas (0.25.1), and Bio (0.1.0) packages. Phylogenetic analysis was based on the concatenation of SNPs using MrBayes software (v3.2.6) and a GTR+G evolution model.
2.4 Cell culture
C-33A (HPV-negative cervical cancer cells), Ca Ski (cervical cancer cells containing 400–600 integrated copies of HPV16 genome), SiHa (cervical cancer cells with 1–2 integrated copies of HPV16 genome), W12 cells (derived from a cervical LSIL with about 100 episomal copies of HPV16 genome per cell), and HeLa cells (cells isolated from an adenocarcinoma with integrated HPV18) were used as negative or positive controls. Cells were cultured in Eagle's minimum essential medium (for C-33A, Ca Ski, and SiHa), Roswell Park Memorial Institute medium (for HeLa), or F-medium (for W12) supplemented with 10% of fetal bovine serum and 1% of streptomycin/penicillin as previously described.12, 13
3 RESULTS
3.1 Patients and sample description
Twelve women, exhibiting a persistent HPV16 infection leading to the development of an HSIL, were included. Each of them had a first HPV16+ cervical sample (P0) at the beginning of the follow-up (with normal cytology or with a mild cytological dysplasia) and a subsequent sample (P2) at the time of a detectable cervical high-grade lesion. The second group of 22 women who spontaneously cleared their HPV16 infections was included. Among them, 13 had a unique HPV16+ detectable sample before clearance (named C2) and 9 had two subsequent HPV16+ samples performed before clearance (named C0 and C2, respectively). Characteristics of these groups are mentioned in Table 1. The median interval between samples P0 and P2, and between C0 and C2 were 36.5 months (range: 6–79) and 16 months (range: 5–31), respectively. HPV16 median viral loads for the 12 P2 and the 22 C2 samples were 6.88 log10 copies/ml (range: 5.74–8.35 log10 copies/ml) and 5.35 log10 copies/ml (range: 3.38–7.57 log10 copies/ml), respectively (p = 0.14).
| Groups | Persistence P (n = 12) | Clearance C (n = 22) | p Value | ||
|---|---|---|---|---|---|
| Mean age at sampling | 29 (min: 21, max: 39) | 31.5 (min: 23, max: 63) | 0.187 | ||
| Tobacco | n = 5 (41.6%) | n = 8 (36.3%) | |||
| Parity | |||||
| Unknown | – | n = 8 (36.3%) | – | ||
| 0 | n = 6 (50%) | n = 6 (27.2%) | 0.265 | ||
| 1 | n = 2 (16.6%) | n = 3 (13.6%) | 1 | ||
| 2–3 | n = 4 (33.3%) | n = 4 (18.1%) | 0.409 | ||
| >4 | – | n = 1 (4.5%) | – | ||
| Contraception at sampling | |||||
| Unknown | n = 1 (8.3%) | n = 2 (9%) | – | ||
| Intrauterine device | n = 1 (8.3%) | n = 1 (4.5%) | 1 | ||
| Oestroprogestogen pill | n = 7 (58.3%) | n = 7 (31.8%) | 0.163 | ||
| Progestogen | – | n = 3 (13.6%) | 0.536 | ||
| Menopause | – | n = 3 (13.6%) | 0.536 | ||
| Local or mechanical | n = 3 (25%) | n = 6 (27.2%) | 1 | ||
| P0 (n = 12) | P2 (n = 12) | C0 (n = 9) | C2 (n = 22) | ||
|---|---|---|---|---|---|
| Cytological status | |||||
| Normal | n = 9 (75.00%) | – | n = 8 (88.89%) | n = 22 (100%) | – |
| ASC-US | n = 1 (8.33%) | – | – | – | – |
| LSIL | n = 2 (16.67%) | – | n = 1 (11.11%) | – | – |
| ASC-H | – | n = 1 (8.33%) | – | – | – |
| AGC | – | n = 1 (8.33%) | – | – | – |
| HSIL | – | n = 10 (83.33%) | – | – | – |
- Abbreviations: AGC, atypical glandular cells; ASC-US, atypical squamous cells of undetermined significance; HPV, human papillomavirus; HSIL, high-grade squamous intraepithelial lesion; LSIL, low-grade squamous intraepithelial lesion.
3.2 Sequencing results
To identify HPV16 genetic variations that may correlate with viral clearance or persistence, whole viral genomes from P0 (n = 12), P2 (n = 12), C0 (n = 9), and C2 (n = 22) samples were sequenced. An unrooted tree of the paired samples C0/C2 and P0/P2 (that excluded the 13 “unique” C2 samples) is shown in Supporting Information: Figure S2 (the Figure also includes the HPV16 genomes from Ca Ski, SiHa, and W12 cells, sequenced as positive controls). Interestingly, all pairs of samples were gathered in the phylogenetic tree, suggesting a low genetic variability within the host compared to the genetic interhost diversity. At the lineage level, 90.5% (n = 19) of the paired genomes belonged to lineage A (including 71.5% in sublineage A1, and 19% in A2), 4.7% (n = 1) to lineage B, and 4.7% (n = 1) to C.
By comparing each sequence with the HPV16 reference genome (GenBank: K02718.1, sublineage A1), we listed and mapped the occurrence of all deletions, insertions, and substitutions inside the whole HPV genomes (Supporting Information: Table S1). Interestingly, most of the genetic deletions and insertions were observed within the intergenic regions between E2 and E5, and E5 and L2 genes (11 different deletions and 14 different insertions), whereas no deletion or insertion was detectable in E4, E6, E7, and L2 genes. When restricting the analysis to subgroups C2 and P2, we failed to detect any significantly more frequent deletion or insertion in either of the two subgroups.
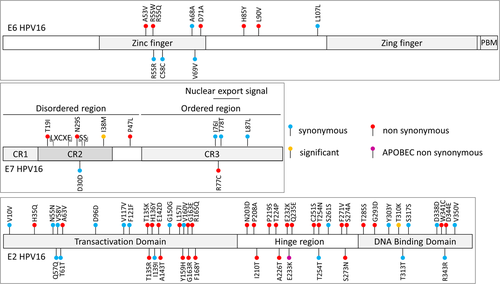
A total of 301 different substitutions was observed (Supporting Information: Table S1). Most of them occurred in the capsid coding genes L1 and L2 (n = 34 and n = 67) or in E1 and E2 (n = 44 and n = 46). Few mutations were observed in the E6 and E7 genes (n = 10 and n = 9). When adjusting to each gene length, E6 remained the least mutated gene (Supporting Information: Figure S3). Among the 301 substitutions (including C2 not paired), 45.8% and 54.2% were observed in HPV genomes belonging to lineages A (n = 138) and B/C/D (n = 163), respectively. The most frequent types of substitutions were the four nucleotide transitions (C>T, T>C, G>A, A>G; 60.9%) compared to the eight nucleotide transversions (all other combinations; 39.1%) (Supporting Information: Figure S4). A total of 23 of the 301 substitutions (7.6%) were compatible with APOBEC3 cytidine deaminase-induced mutations since they occurred in a TCW motif (where W is A or T) but none were observed in E5, E6, and E7 genes even though there were compatible motif available on their sequence (Supporting Information: Figure S5). Due to the importance of E6, E7, and E2 genes in HPV-induced carcinogenicity, substitutions in these genes are specifically shown in Figure 1. When comparing the occurrence of nonsynonymous substitutions in HPV16 from the C2 and P2 subgroups, only one mutation, occurring in L2 (L330F) was significantly more frequent in samples from the subgroup C2 than in P2 (n = 18 [81.8%] and n = 5 [41.6%], respectively, p = 0.025). The seven other significant differences in frequency between the subgroups are presented in Supporting Information: Table S2, including one mutation in E7 (I38M, more frequent in P0 than in C2; p = 0.01), but no mutation in E6.
4 DISCUSSION
The prognosis of cervical infection by an hr HPV depends on the viral genotype,14 lineage, sublineage, or even variant involved.6 Identifying these specific variants and/or the molecular determinants linked to these variants, which may either favor viral persistence (by hijacking the immune system, in particular) or promote the development of cancers through a more carcinogenic phenotype, is still challenging. Moreover, the distribution of variants worldwide is influenced by geographic and ethnic factors.15
Here, we aimed to identify specific HPV16 genetic variations associated with viral persistence and carcinogenicity. For that purpose, serial samples from HPV16-infected women were sequenced by NGS. By extensively exploring the occurrence of genetic deletions, insertions, and substitutions inside the HPV16 whole genomes, we failed to detect any significant molecular variations associated with viral clearance or the development of (pre-) cancerous lesions associated with viral persistence, with the exception of the mutation L330F which was significantly more frequent in women who had cleared their HPV16 infection (P2 vs. C2 subgroups). Inside the HPV16 genomes, we observed a high degree of conservation in the E6 and E7 oncogenes, since we did not observe any deletion or insertion in these genes, and only a few base substitutions, without any APOBEC3-compatible substitution.
Phylogenetic analysis of HPV16 sequences obtained longitudinally in our cohort revealed high within-host conservation, compared to the between-host diversity. Other groups have observed high diversity of HPV16 genomes between women, contrasting with a higher degree of conservation in HPV16 genomes obtained at multiple body sites in a unique woman16 or obtained longitudinally in patients harboring a persistent infection.17
As observed in other cohorts, the genetic conservation of HPV16 was not homogeneous along the genome.16, 17 Deletions and insertions predominated in noncoding regions, as observed in another study,17 whereas the oncogenes E6 and E7 exhibited a high degree of conservation. In a large cohort of HPV16-infected women, Mirabello et al.16 have described higher conservation of E7 in HPV16 variants associated with (pre-)cancers than in HPV16 variants from the control group and suggested that this conservation was critical for HPV16 carcinogenesis by disrupting the pRb function. In our study, the I38M substitution located in the CR2 region of E7 was more frequently observed in the subgroup P0 than in C2 (p = 0.10) although this mutation disappeared between the first (P0) and the last (P2) samples in three patients. Phenotypical analyzes could be performed to investigate the possible carcinogenic impact of this mutation. The disappearance of this mutation in three patients between P0 and P2 may suggest either a selective pressure that had led to its elimination or a possible action of this mutation through a “hit and run mechanism” at the beginning of infection (P0). Alternatively, this apparent elimination of the mutation in three patients may correspond to a multiple infections by two HPV16 variants with one overgrowing the other between P0 and P2.
Interestingly, no APOBEC3-compatible substitution was observed in E5, E6, and E7 in our study, contrasting with a high rate of APOPEC3-compatible substitutions in E2 and L2. Similar variations of APOBEC3-induced mutations across the HPV16 genome were also recently described in another cohort.18 In this latter study, the authors observed a significantly reduced number of APOBEC3 mutations in HPV16 genomes during (pre-) cancers compared to controls.
The L330F mutation in the minor capsid L2 protein was the only significant genetic variation observed between the C2 and the P2 subgroups of patients. The protein L2 plays major roles in host cell entry, disruption of endosomal membranes, subcellular trafficking of the viral genome, and its encapsidation.19, 20 Since this mutation is located in the central region of the sequence, far from the important DNA binding domains, the furin cleavage site, and the identified neutralizing epitope regions (all located at the N-terminal part of the protein), its biological relevance is unclear. The prevalence of this mutation among HPV16 sequences from different lineages is unknown but its presence has been recently observed in 43.5% of HPV16-positive specimens, mainly classified in lineage A, for which the L2 sequence variations were investigated.21 While the clinical impact of this mutation was not explored in this latter study, a previous case report had identified this mutation in all HPV16 sequences obtained after the microdissection of a cervical invasive squamous cell carcinoma coexisting with multiple CIN2 and CIN3 lesions in a unique patient.22 Further studies are required to explore the phenotypic impact of this mutation or to better characterize a possible polymorphism in this position among the L2 sequence.
Our study has some limitations. First of all, only a small number of samples were available in each subgroup and most of them belonged to the HPV16 lineage A. A longitudinal follow-up of HPV16 variations could be performed in a larger cohort of women, infected by different HPV16 (sub-)lineages, to complete our observations. Large cohorts should also distinguish between the occurrence risk of HSIL and cervical cancers.6 Exploring the phenotypic impact of the main genetic variations observed in our study is required to determine their interest in clinical practice.
In conclusion, longitudinal analysis of HPV16 whole genomes from a cohort of HPV16-infected women revealed a low genetic intrahost variability compared to the interhost diversity, and no correlation between genetic variations and occurrence of precancerous lesions.
AUTHOR CONTRIBUTIONS
Conceptualization, methodology, investigation: Alice Debernardi. Methodology, software, and formal analysis: Benoit Valot, Julien Almarcha, and Didier Hocquet. Conceptualization, methodology: David Guenat. Resources: Marie-Paule Algros, Didier Riethmuller, and Rajeev Ramanah. Visualization and supervision: Christiane Mougin. Conceptualization, methodology, project administration, funding acquisition, supervision: Jean-Luc Prétet. Conceptualization, methodology, investigation, writing – original draft, supervision: Quentin Lepiller.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Gilles Travé for his scientific collaboration and advice regarding the structural biology of human papillomavirus proteins.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.