The emergence of Omicron lineages BA.4 and BA.5, and the global spreading trend
The Omicron BA.4 and BA.5 lineages, which recently emerged in April–May 2022,1-3 have spread to many parts of the world within 2 months. BA.2 in April 2022 almost completely replaced BA.1, which had dominated the Omicron lineages earlier this year (Supporting Information: Figure 1A-D). Furthermore, BA.3 has become less dominant than other lineages (Supporting Information: Figure 1E-F). It is important to note that the BA.4 and BA.5 lineages began to dominate the world from their appearance (April 2022) until the date (mid-June), and around the same time, BA.2 began to decline (Supporting Information: Figure 1C-D,G-J). If this trend continues, we can expect BA.4 and BA.5 lineages to replace BA.2 lineage almost entirely as soon as possible and become more dominant.
As of June 16th, 2022, the BA.4 lineage has been reported from 55 countries with a worldwide prevalence of 8% (June 10th, 2022) (Supporting Information: Figure 1G-H). Furthermore, it is noteworthy that 69.31% of the BA.4 sequences detected up to June 16th, 2022 were from three countries: South Africa (31.24%), the United States (27.05%), and the United Kingdom (11.02%) (Figure 1A). As with BA.4, more than 80% of the BA.5 sequences detected up to June 6th, 2022 are found in the United States (33.25%), South Africa, and European countries (Figure 1B). However, it should be noted that the BA.5 lineages have spread to 54 countries and have an 11% (June 10th, 2022) prevalence worldwide as of June 16th, 2022 (Supporting Information: Figure 1I-J). Interestingly, more than 80% of the BA.5.1 sequences detected up to June 6th, 2022 are found only in European countries (Figure 1C) and have a global prevalence of 8% (June 10th, 2022) (Supporting Information: Figure 2A-B). Curiously, as of June 16th, 2022, the BA.4 and BA.5 lineages have replaced the BA.2 lineages in South Africa, with the BA.4 and BA.5 lineages becoming the dominant lineages (Figure 1D). Furthermore, it is revealed that the BA.4 lineages have become the dominant lineages in the BA.4 and BA.5 lineages in South Africa (Figure 1E). Remarkably, there are a total of four amino acid differences between the BA.4 and BA.5 lineages, in proteins such as OFR1a, ORF6, ORF7b, and Nucleocapsid (N), with one amino acid difference in each protein; and did not express any amino acid differences in proteins such as ORF1b, Spike, Envelope (E), Membrane (M), ORF8, and ORF10 (Figure 1F–O). Furthermore, it is important to note that only one amino acid in ORF10 differs between the BA.5 and BA.5.1 sub-lineages (Figure 1O). Similarly, OFR1a-DEL141/143, ORF7b-D61L, and N-P151S mutations are specific to BA.4, and ORF6-D3N mutation is specific to BA.5/BA.5.1 lineage (Figure 1F–O).
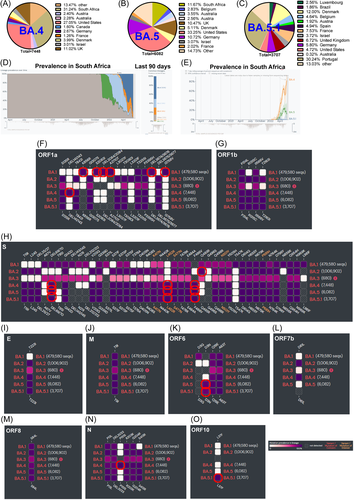
Our previous study reported differences in the spike protein between the BA.1, BA.2, and BA.3 lineages4; accordingly, the BA.4 and BA.5 lineages are the closest to the BA.2 lineage in the spike protein. There are only four amino acid levels in Spike protein that differ between the BA.4/BA.5 and BA.2 lineages. Of these four mutations, mutations such as DEL69/70 and L452R are already present in the Omicron lineage B.1.1.529,5 and L452R was also detected in the delta (B.1.617.2) variant, reported affecting the vaccine-induced neutralizing antibodies activity.6 Furthermore, the Q493R (spike protein) mutation was found in all Omicron lineages, and sublineages are not found in the BA.4/BA.5 lineages (Figure 1H). On the other hand, the spike-F486V mutation is found only in the BA.4/BA.5 Omicron lineages (Figure 1H). Interestingly, the spike-F486V mutation has been accumulating spike protein since the beginning of this year (https://cov-spectrum.org/explore/World/AllSamples/AllTimes/variants?aaMutations=S%3AF486V&). In particular, F486L mutation has been reported to increase virus entry into mink/ferret ACE2 expressing cells,7 and mutation at F486 decreases neutralizing activity of neutralizing antibodies.1 BA.4/BA.5 has been found to be 4–20 times more resistant to monoclonal antibodies such as cilgavimab and Evusheld than BA.2.6, 8 Remarkably, it is important to note that BA.4/BA.5 lineages evade the postvaccination (3-dose) BA.1 infections mediated antibodies neutralization.8 For these reasons, it is possible to speculate that the BA.4/BA.5 lineages are spreading very fast worldwide. However, since the spike protein is almost identical in the BA.2 and BA.4/BA.5 lineages, antibodies to the BA.2 infection can be expected to protect the BA.4/BA.5 lineages. Furthermore, vaccines that use the Omicron (BA.2) strain/spike protein can be expected to control the spread and protect against BA.4/BA.5 lineages.
In India, it was 2338 new cases/Day on May 30th, 2022 and has been increasing to 13 216 new cases/Day on 17th June 2022 within 3 weeks (https://outbreakindia.com/india-dashboard), and a similar situation exists in many other countries. Correspondingly, in India, during these periods, the BA.2 prevalence began to decline, and the BA.4 prevalence increased (Supporting Information: Figure 2C). From these, it can be inferred that the BA.4 lineage is causing the increasing number of COVID-19 cases in India. Furthermore, it should be noted that there are no major reports of BA.4/BA.5 lineages causing severe illness. However, large-scale research into the severity of the disease in the BA.4/BA.5 lineages is required. Finally, the BA.4/BA.5 lineages are expected to spread rapidly in India and other countries, causing the next COVID-19 wave shortly. Therefore, adherence to the correct COVID-19 protocol and vaccination strategy to protect against BA.4/BA.5 lineages seems essential.
AUTHOR CONTRIBUTIONS
All the authors contributed significantly to this manuscript. Perumal A. Desingu analyzed and wrote the first draft, and Kumaresan Nagarajan reviewed the manuscript. All the authors reviewed and approved the final submission.
ACKNOWLEDGMENTS
P.A.D. is a DST-INSPIRE faculty supported by the Department of Science and Technology (DST/INSPIRE/04/2016/001067), the Government of India, and the Science and Engineering Research Board (SERB) (CRG/2018/002192), Government of India.
CONFLICT OF INTEREST
The author declares no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
Not applicable.