TRIM22 inhibits respiratory syncytial virus replication by targeting JAK-STAT1/2 signaling
Abstract
Respiratory syncytial virus (RSV) infection is a major cause of lower respiratory tract disease. Although RSV causes major economic losses every year, effective treatments have not been found so far. Recent studies have shown that the tripartite motif-containing (TRIM) superfamily plays an essential role in the immune response. In this study, we found that TRIM22 had an inhibitory effect on RSV infection, and downregulation of TRIM22 moderately enhanced RSV replication. Our data further demonstrated that RSV infection induced TRIM22 expression through the activation of JAK-STAT1/2 signaling. RSV infection also induced TRIM22 expression. Taken together, these data points showed that the TRIM family member, TRIM22, had an essential role in resisting RSV infection, and this effect was closely related to the JAK-STAT1/2 pathway. Our results provide promising evidence for a novel target for the prevention and treatment of RSV.
Highlights
-
Knockdown of TRIM22 promoted the proliferation of RSV in cells.
-
The expression of TRIM22 was regulated by the JAK-STAT1/2 signaling pathway.
-
Interferon resistance to RSV infection depended on TRIM22.
-
RSV infection activated the JAK-STAT1/2 signaling pathway and induced the expression of TRIM22.
1 INTRODUCTION
Respiratory syncytial virus (RSV) is widely recognized as the most significant pathogen responsible for viral acute lower respiratory tract illness in infants and young children.1 Approximately 34 million children are infected with RSV each year, with approximately 10% requiring hospitalization. Of the total number of deaths resulting from RSV, 99%, or as many as 200 000, are contributed by developing countries.2 A sample of 56 569 children in Argentina showed that 1239 children had respiratory infections, and 61.6% of these were caused by RSV.3 In the United States, a large proportion of outpatients under the age of 5 are infected with RSV.4 In addition, the target population for RSV infection includes both elderly and immunosuppressed individuals. For immunocompromised patients, especially those with hematological malignancies and human stem cell transplant recipients, the risk of RSV-related diseases is significantly increased.5
RSV is an enveloped virus with a negative-sense RNA genome. The single-stranded, nonsegmented 15.2 kb RSV genome consists of 10 genes that encode 11 proteins.6 Three structural proteins exist in the RSV envelope: glycoprotein (G), fusion glycoprotein (F), and small hydrophobic protein (SH). In theory, the infection model of RSV is similar to the infection model of other enveloped viruses, in which the G protein begins to combine with host cells by interacting with glycosaminoglycans (GAGs),7, 8 and then the F protein mediates the merging of the host membrane with the virus envelope to allow the virus to enter the cell.9 For decades, there has been a heightened awareness of the major socioeconomic burden caused by RSV-related diseases. However, targeted therapy has not been recommended until now, and there are no specific treatments currently used in the clinic.10, 11 However, an increasing number of studies are being performed to develop antiviral therapies targeting RSV and antiviral drugs for commercial use.
Tripartite motif containing 22 (TRIM22) has been shown to inhibit various viruses. For instance, its ubiquitination can inhibit encephalomyocarditis virus (ECMV) replication, resulting in antiviral effects.12 Furthermore, hepatitis B virus (HBV) replication can also be inhibited by TRIM22, via interference with the HBV core promoter.13 Recent studies have shown that the E3 ubiquitin ligase activity of TRIM22 is a crucial factor in the degradation of the influenza A virus (IAV) nucleoprotein, which is indispensable for viral replication,14 and the human hepatitis C virus NS5A protein.15 It has also been shown that TRIM22 is an interferon-inducible p53 target gene that plays an important role in cell proliferation.16
Increasing evidence supports the antiviral effects of TRIM22. However, its effect on RSV replication remains unknown. In this study, the effect of TRIM22 on RSV replication in vitro was explored. RSV replication was shown to increase when TRIM22 expression was silenced, indicating that TRIM22 inhibits RSV replication.
In this study, we showed that RSV replication was increased by the knockdown of Janus kinase 1 (JAK1) in A549 and HEP-2 cells. Although RSV infection did not affect JAK1 expression levels, it was shown to activate the JAK-STAT1/2 signaling pathway.
2 MATERIALS AND METHODS
2.1 Materials
Antibodies against JAK1 (Cat# 29261), p-JAK1 (Cat# 74129), PKC (Cat# 2056), and Actin (Cat# 3700) were purchased from Cell Signaling Technology (Beverley, MA). The antibody against TRIM22 (Cat# HPA003307) was purchased from Sigma-Aldrich. Recombinant Human IFN-γ Protein (Cat# 285-IF) and JAK1 inhibitor I (Cat# 6577) were obtained from R&D Systems company. shRNA expression vector (Cat# C02003), negative control shRNA expression vector (Cat# C03002), and oligonucleotides were obtained from GenePharma company. The sequences of oligonucleotides are summarized in Table S1.
2.2 Cell culture
A549 and HPE-2 cells were cultured in Dulbecco's Modified Eagle's medium (DMEM) complete medium (Cat# 11965-092; Gibco) containing 10% fetal bovine serum, penicillin (100 U/mL), and streptomycin (100 μg/mL). Human pulmonary microvascular endothelial cell (HPMEC) was cultured in ECM complete medium (Cat# 1001; ScienCell), containing 5% fetal bovine serum, penicillin (100 U/mL), and streptomycin (100 μg/mL). Cells were cultured at 37°C in a humidified incubator with 5% CO2.
2.3 RSV infection
RSV strain A2 was propagated in HEP-2 cells. HEP-2 cells were inoculated with RSV at a multiplicity of infection of 0.01 per cell and incubated at 37°C for 2 hours. The inoculum was then removed and fresh complete DMEM was added. Viruses were harvested 4 to 5 days post-infection. The culture medium was collected, centrifuged at 2000 rpm for 20 minutes at 4°C, and filtered through a 0.22 μm filtration system (Cat# SLGP033RB; Millipore). Aliquots were frozen at −80°C. The RSV titer was determined using a 50% tissue culture infectious dose (TCID50) assay. RSV was titrated using the Reed-Muench method, to calculate the TCID50, which was then converted to plaque-forming units using a conversion factor.
2.4 Cell transfection
To suppress gene expression, cells were transfected with small interfering RNAs (siRNAs). All siRNAs were transfected into cells at the indicated concentrations using Lipofectamine 3000 (Cat# L3000001; Invitrogen), according to the manufacturer's protocol. Silencing efficiency was determined by quantitative polymerase chain reaction (qPCR) and Western blot analysis.
2.5 Quantitative reverse transcription PCR
Total RNA was isolated using an RNeasy Mini Kit (Cat# 74104; Qiagen) according to the manufacturer's instructions. Reverse transcription (RT) was performed using 1 μg of denatured RNA and 100 pmol of random hexamers (Cat# N8080127; Invitrogen), in a total volume of 25 μL containing 12.5 U MultiScribe reverse transcriptase (Cat# 4311235; Invitrogen), as previously described.17 Quantitative RT-PCR (qRT-PCR) analysis was performed to measure relative messenger RNA (mRNA) levels using an SYBR Green kit (Cat# 1708891; Bio-Rad Laboratories Inc). Samples were normalized to actin expression levels. The sequences of the primers used for qRT-PCR are presented in Table S1.
2.6 Western blot analyses
Briefly, protein extracts were fractionated on 10% SDS-PAGE gels and transferred onto hydrophobic PVDF transfer membranes (Cat# IPVH00010; Millipore). The membranes were blocked with 5% nonfat milk in Tris-buffered saline/Tween 20 buffer, and probed with primary antibodies. Following incubation with horseradish peroxidase-conjugated secondary antibodies, the proteins were visualized using a Western blot detection kit (Cat# RPN2106; GE Healthcare).
2.7 Stable transfection of shRNA plasmids
A TRIM22-specific short hairpin RNA or small hairpin RNA (shRNA) was cloned into the pGPU6/Neo vector and its insertion was confirmed by Sanger sequencing. A549 cells were transfected with the pGPU6/Neo-TRIM22 shRNA plasmid vector. Selection was performed using 400 μg/mL neomycin (G418) and stable clones were maintained in 100 μg/mL neomycin. Silencing of the target gene was confirmed by Western blot analysis.
2.8 Statistical analyses
Investigators were not blinded to the allocation during the experiments or to the outcome assessment. Descriptive statistics are provided in the figure legends. Given the nature of the experiments and the types of samples, differences in continuous variables were assessed using the nonparametric Mann-Whitney test. All data were analyzed using GraphPad Prism statistical software.
3 RESULTS
3.1 TRIM22 inhibited RSV infection
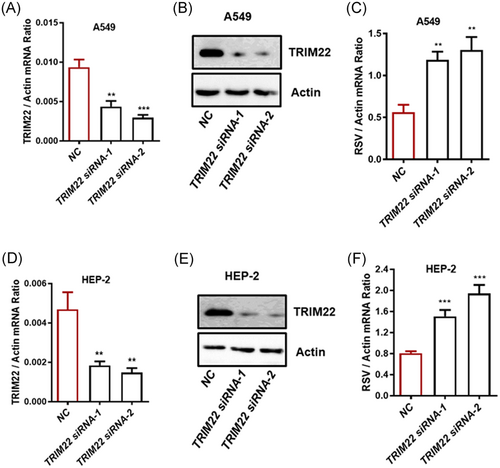
A549 is a human non–small-cell lung cancer cell line and HEP-2 is a laryngeal squamous cell carcinoma cell line. Both cell lines are sensitive to RSV infection.18, 19 To investigate the functional role of TRIM22 in RSV replication, we generated two siRNAs against TRIM22. The TRIM22 gene was silenced by siRNA in A549 cells. Compared to their levels in control cells, TRIM22 mRNA and protein levels were consistently reduced in TRIM22-knockdown cells (Figure 1A,B) infected with RSV. The viral burden of these cells was assessed by qPCR post-infection. TRIM22 knockdown moderately increased the RSV load in A549 cells (Figure 1C). We next investigated the antiviral role of TRIM22 in HEP-2 cells. The knockdown of TRIM22 consistently resulted in a higher RSV load in these cells (Figure 1D-F). To investigate the antiviral role of TRIM22 in primary cells, we infected HPMECs with RSV after silencing TRIM22 (Figure S1A). RSV replication increased moderately after siRNA-mediated TRIM22 knockdown (Figure S1B). Taken together, these results indicated that TRIM22 suppresses RSV replication.
3.2 TRIM22 expression was regulated by JAK-STAT1/2 signaling
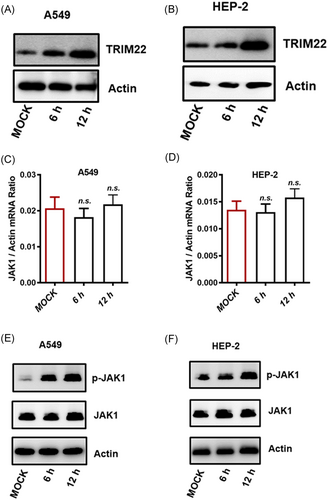
Recent studies have shown that the tyrosine kinase, JAK-STAT1/2, plays an indispensable role in the induction of TRIM22 expression. The inhibition of protein kinase C (PKC) activity also significantly attenuated the induction of TRIM22 expression by interferon gamma (IFN-γ). It has also been reported that the overexpression of JAK1 and PKC activates TRIM22 promoter activity, and the inhibition of the JAK-STAT1/2 and PC-PLC/PKC pathways significantly attenuates IFN-γ-induced IRF-1 expression in HepG2 cells.20 To determine if TRIM22 is a functional target of JAK-STAT1/2 in A549 and HEP-2 cells, siRNA knockdown of JAK1 was performed in A549 and HEP-2 cells. JAK1 protein levels were reduced in A549 and HEP-2 cells after JAK1 knockdown (Figure 2A,D). JAK1 knockdown resulted in the downregulation of TRIM22 expression in A549 (Figure 2B,C) and HEP-2 cells (Figure 2E,F). We next determined the role of PKC in the regulation of TRIM22 expression. PKC knockdown significantly decreased the expression levels of PKC protein in A549 (Figure S2A) and HEP-2 cells (Figure S2C). However, no difference was observed in TRIM22 RNA levels between the PKC knockdown and control groups (Figures S2B and S2D). Thus, we concluded that TRIM22 was a direct target of JAK1 in A549 and HEP-2 cells. These results indicated that the expression of TRIM22 in A549 and HEP-2 cells was regulated by the JAK-STAT1/2 pathway, rather than the PC-PLC/PKC pathway.

The results described above showed that TRIM22 expression was regulated by JAK-STAT1/2 signaling. Therefore, we performed loss-of-function assays in A549 and HEP-2 cells to assess whether JAK1 influences RSV replication. JAK1 downregulation led to a moderate increase in RSV replication in A549 (Figure 3A) and HEP-2 cells (Figure 3B). TRIM22 expression has been shown to be modulated by IFN-γ induction.20 Thus, to assess whether the antiviral effect of JAK-STAT1/2 signaling was dependent on TRIM22, we generated a TRIM22 shRNA cell line (Figure 3C). A549 cells were infected with RSV and when the infected cells were treated with IFN-γ, viral load significantly decreased. This effect was inhibited in the TRIM22 shRNA cell line (Figure 3D). The data presented above showed that JAK1 also played an important antiviral role. To further investigate the role of JAK1, we treated A549 cells with JAK1 inhibitor I, a pan JAK1 inhibitor. Viral replication was enhanced in A549 cells treated with JAK1 inhibitor I, but this effect was inhibited in the TRIM22 shRNA cell line (Figure 3E). Taken together, these results showed that the antiviral role of JAK-STAT1/2 signaling was dependent on TRIM22.

3.3 Activation of the JAK-STAT1/2 signaling pathway by RSV infection
To further investigate whether JAK-STAT1/2 signaling affects RSV replication, A549 and HEP-2 cells were infected with RSV. TRIM22 mRNA and protein expression levels were determined in A549 and HEP-2 cells. As expected, RSV infection moderately increased the expression levels of TRIM22 in both A549 and HEP-2 cells (Figure 4A,B and Figure S3A,B). However, no changes were observed in the expression levels of JAK1 following RSV infection in either A549 or HEP-2 cells (Figure 4C,D). Additionally, Western blot analysis of protein extracts from cells infected with RSV showed that the level of phosphorylated JAK1 (p-JAK1) was increased in A549 and HEP-2 cells. However, total JAK1 expression levels were not altered (Figure 4E,F). Collectively, these data showed that RSV infection promoted JAK1 phosphorylation and activated IFN signaling and that RSV replication was inhibited by regulating TRIM22 expression.
4 DISCUSSION
Lower respiratory tract infections caused by RSV are prevalent worldwide, mainly targeting children and the elderly. Although RSV has been extensively studied, there is currently no effective treatment, except for passive immunity.21 Our results showed that RSV infection was inhibited by TRIM22.
TRIM22, also known as staf50, belongs to the C-IV group in the tripartite motif-containing (TRIM) family.22-24 The antiviral activity of TRIM22 against different viruses has previously been proven. TRIM22 has a typical RBCC structure, including a RING domain with E3 ubiquitin ligase activity.25 In this motif, cysteine-15 and cysteine-18 constitute the catalytic site that mediates the transfer of ubiquitin to the target proteins.12, 26 The TRIM22 RING domain promotes a self-ubiquitination pathway that leads to proteasomal degradation and is also related to virus recognition.12, 27 The B domain (B-box 2) of TRIM22, which is essential for nuclear localization, is located downstream of the RING domain.28 Moreover, a CC domain with an unclear biological function and a PRYSPRY domain at the C-terminus are also found in TRIM22.29 Specifically, the PRYSPRY domain, known as B30.2, is composed of three hypervariable regions, and it is crucial for anti-HIV activity.30 The replication of the porcine reproductive respiratory syndrome virus (PRRSV) in MARC-145 cells has been shown to increase after TRIM22-RNA interference treatment. However, the ability of TRIM22 to inhibit PRRSV replication is lost when the SPRY domain or nuclear localization signal (NLS) is deleted. Co-immunoprecipitation analysis has shown that the NLS motifs of both the N protein and the PRRSV nucleocapsid protein are involved in the interaction with TRIM22, and the latter is processed through the SPRY domain.31
Previous investigations have shown that TRIM22 plays a critical role in IFN-mediated antiviral activity. TRIM22 expression can be induced by IFN-γ, through the activation of JAK-STAT1/2 and the PC-PLC/PKC signaling pathway.20 In the present study, TRIM22 expression was inhibited by JAK1 knockdown, but not PKC knockdown, in A549 and HEP-2 cells. This result indicated that the regulation of TRIM22 expression through JAK-STAT1/2 signaling was independent of PKC. It also suggested that the inhibition of RSV replication by JAK-STAT1/2 occurred partially through TRIM22. Further studies are under consideration to confirm this finding.
We also measured TRIM22 expression after RSV infection. We observed that TRIM22 was upregulated in RSV-infected A549 and HEP-2 cells, but JAK1 expression was not affected. Thus, JAK1 may have inhibited RSV replication partially through TRIM22. The finding that RSV replication activated the JAK-STAT1/2 signaling pathway was not consistent with previous reports that PRRSV suppresses the JAK1-STAT3 pathway.32, 33
In this study, we present evidence highlighting the importance of JAK-STAT1/2 signaling in regulating RSV pathogenesis. However, the mechanisms by which JAK-STAT1/2 signaling modulates RSV replication remain unclear.
ACKNOWLEDGMENTS
This study was funded by grants from the National Natural Science Foundation of China (31700148), and Shenzhen San-Ming Project for prevention and research on vector-borne diseases (SZSM201611064).
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
XX designed the experiments and YW wrote the manuscript. XX and YC performed the majority of the experiments and analyzed data. YL and YQ helped with the RNA isolation and qPCR detection. RZ contributed experimental suggestions and strengthened the writing of the manuscript. All authors reviewed, critiqued, and provided comments to the text.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of these studies are available from the corresponding author upon request.