Respiratory syncytial virus in influenza-like illness cases: Epidemiology and molecular analyses of four consecutive winter seasons (2014-2015/2017-2018) in Lombardy (Northern Italy)
Laura Pellegrinelli and Cristina Galli contributed equally to the study.
Abstract
Background
Besides seasonal influenza viruses (IV), several other pathogens—including respiratory syncytial virus (RSV)—are involved in clinically undistinguished influenza-like illnesses (ILIs). This study aimed at investigating the contribution of RSV in ILI cases in Lombardy (Northern Italy) during four consecutive winter seasons.
Materials and Methods
In the framework of influenza surveillance, respiratory samples from ILI outpatients were collected from 2014-2015 to 2017-2018 season. IV-negative swabs were included in the study and analyzed to detect and molecularly characterize RSV-A and RSV-B.
Results
A total of 12.9% (135/1047) of samples were positive to RSV that was mostly detected among children ≤5 years (51/183, 27.8%) and those aged 6 to 15 years (30/158, 18.9%), whereas elderly >65 years accounted for 12% of RSV cases (15/125). The median start of RSV epidemic was in the end of November, with a peak in mid-February and a width of nearly 4 months, almost overlapping seasonal influenza epidemic. RSV-A and RSV-B co-circulated in all considered seasons, with RSV-B predominating on RSV-A (63.6% vs 36.4%; P < .001). Most (85.2%) RSV-A belonged to genotype ON1 and the remaining to NA1. All RSV-B clustered within the BA genotype.
Conclusions
In this study, RSV significantly contributed to ILI cases, especially among pediatric population (<15 years), although it was detected in all age groups. RSV-B predominated on RSV-A, and the most recent evolved genotypes (BA and ON1, respectively) circulated. Investigating the epidemiological and molecular characteristics of RSV in ILI cases can increase baseline epidemiological information before the introduction of RSV vaccination.
Highlights
-
RSV significantly contributed to influenza-like illnesses (ILIs) cases, especially among pediatric population <15 years
-
RSV-B predominated on RSV-A and the most recent evolved genotypes BA and ON1 circulated among ILI cases.
-
To set up specific vaccination strategies, it is warrant increasing the current knowledge on RSV epidemiology and impact.
1 INTRODUCTION
Human respiratory syncytial virus (RSV) is a common virological cause of acute respiratory infection, infecting especially children, despite its role in adults and elderly has been increasingly recognized. RSV infection can be asymptomatic or associated with symptoms ranging from mild—such as influenza-like illness (ILI)—to severe and life-threatening illness resulting in severe bronchiolitis or pneumonia, and requiring hospitalization.1 In Europe, RSV shows a seasonal distribution mainly from October to May, with seasonal peaks in January/February, which generally overlap seasonal influenza epidemic.2
RSV is a single-stranded negative-sense RNA virus belonging to the Pneumoviridae family, genus Orthopneumovirus.3 Its genome is composed of about 15 000 nucleotides (nt), encoding for 10 viral proteins3; among them, the surface glycoproteins G and F play a significant role in viral pathogenicity and immune evasion.1, 4 Based on antigenic variability of these surface proteins, RSV is classified into two distinct subgroups (A and B),5 which exhibit a wide genetic diversity—especially in the glycoprotein G gene—that results in divergence into many genotypes.6 RSV-A and RSV-B can circulate simultaneously but usually each season is characterized by the predominance of one subgroup,7 with RSV-A infection leaning toward outweighing and worsening the clinical manifestations than RSV-B,7-9 despite the association between RSV genotypes and disease severity remains unclear.7-10
A number of RSV vaccines are under development11 and, in the perspective to set up specific vaccination strategy, baseline data on RSV circulation and its distribution among the different age groups are essential. At the present time, 15 European countries have already established a laboratory-based RSV surveillance system, to define the burden, the seasonality, and the geographical spread of RSV infection.12 A well-established influenza surveillance system has been recognized as a valuable tool to monitor the circulation and impact of RSV13 and can be used for RSV surveillance based on the current case definition of ILI.14 In Italy, no RSV surveillance system has been implemented so far, and only limited information regarding the molecular epidemiology of RSV are available from individual research studies.7, 8, 15-17
The aim of this study was to assess for the first time the molecular epidemiology of RSV among ILI outpatients, identified in the framework of the influenza surveillance system, in Lombardy from 2014-2015 to 2017-2018 winter season to (a) estimate the RSV positivity rates among ILI cases, and (b) perform the molecular characterization and phylogenetic analysis of circulating RSV.
2 METHODS
2.1 Influenza surveillance network, ILI case definition, and study population
The Italian Influenza Surveillance Network (InfluNet) is coordinated by the Italian Ministry of Health and is based on the voluntary participation of sentinel physicians (pediatricians and general practitioners) who survey about 2% of the general population seeking care in ambulatory facilities for ILI occurrence.18 Sentinel physicians weekly report data on the number of ILI cases and collect respiratory samples (nasal/oropharyngeal swabs) from their outpatients for virological surveillance. The standard case definition of ILI is an abrupt onset of fever (>38°C) or feverishness, one or more respiratory symptoms (cough, sore throat, and/or shortness of breath), and one or more systemic symptoms (myalgia, headache, and/or malaise).14 In Italy, virological surveillance of seasonal influenza starts on week 46 (mid-November) and ends on week 17 (end of April) of the following year.18 For each ILI case, information on age, sex, influenza vaccination status, and presence of underlying medical conditions (such as cardiovascular diseases, chronic respiratory diseases, metabolic diseases, and immunosuppression) are collected anonymously.
All ILI cases occurred in outpatients and recognized by physicians operating in the framework of InfluNet in Lombardy (a region in Northern Italy accounting for nearly 10 million inhabitants equal to about 1/6 of the entire national population) and tested negative to influenza virus (IV) during four consecutive winter seasons (from 2014-2015 to 2017-2018) were included in this study.
2.2 Ethical statement
This study was performed according to the Institutional Review Board guidelines concerning the use of biological specimens for scientific purposes in compliance with Italian law (art.13 D.Lgs 196/2003). Approval from an ethics committee and informed consent for virus detection were not required since data and samples from outpatients with ILI were collected and analyzed anonymously within the National Influenza Surveillance Program.
2.3 Molecular detection of RSV-A and RSV-B
RNA was extracted from 200 μL of each respiratory sample and eluted in 100 μL of elution buffer using the Invisorb® Spin Virus RNA Mini kit (Stratec Biomedical AG, Germany), according to the manufacturer's instructions. To check the extraction performance, a one-step real-time retro-transcription (RT) polymerase chain reaction (PCR) assay targeting human ribonuclease P gene (RNase P) was carried out (TaqMan® RNase P Assay, ABY® dye/QSY® probe; Thermo Fisher Scientific); samples showing a RNase P cycle threshold (Ct) value <35 were considered suitable to be tested for virus detection.
A one-step real-time RT-PCR assay was performed to simultaneously detect IV type A and B using specific primer/probe sets targeting the matrix (M) gene and the nucleoprotein (NP) gene, as reported previously.19 After the confirmation that samples were eligible to be included into the study (RNaseP Ct < 35 and IV-negative), a one-step RT-PCR assay was performed to detect RSV using specific primer/probe set targeting the matrix (M) gene (nt: 3254-3337 of the reference strain, isolate A2; accession number [AN]: NC038235).20 The RSV real-time RT-PCR assay was set up using 5 µL of RNA in a final reaction volume of 20 µL by using Luna Universal Probe One-Step RT-qPCR kit (New England Biolabs Inc), in a StepOne Plus real-time PCR System (Thermo Fisher Scientific). A sample was considered positive when its Ct value was less than 39.
RSV-positive samples were then tested to discriminate between subgroup A and B by a RT-multiplex-nested-PCR assay by using sets of primers targeting the F gene of RSV-A (nt: 6008-6371 of the reference strain, isolate A2; AN: NC038235) and RSV-B (nt 5696-6307 of the reference strain, isolate B1; AN: AF013254).21 Following RT with random primers, 10 µL of complementary DNA was amplified in a final reaction volume of 50 µL, then 5 µL of amplified material was nested by using GoTaq® G2 DNA Polymerase (Promega Corporation) in a GeneAmp® PCR System 9700 (Thermo Fisher Scientific).
2.4 Sequencing and molecular characterization of RSV-A and RSV-B
Approximately 50% of RSV-A- and RSV-B-positive samples were randomly selected from each season and sequenced. Sequences were obtained directly from clinical specimens by a RT-nested-PCR assay. A fragment of 988 nt encompassing the entire G gene of RSV-A and RSV-B (nt: 4682-5669 of the reference strain, isolate A2; AN: NC038235) was amplified by using GoTaq® G2 DNA Polymerase (Promega Corporation) and specific primers.22 The amplicons were purified by using a commercial kit (NucleoSpin Gel® and PCR clean-up kit; Macherey-Nagel, Germany) and sequenced by Sanger method.
The sequences included in this study were edited and assembled by using BioEdit software23; all study sequences were submitted to the GenBank® database on the the National Center for Biotechnology Information (NCBI) website24. The study sequences were aligned with reference strains of previously published genotypes retrieved from GenBank® database on the NCBI website24 by using the ClustalW program and implemented in the alignment editor BioEdit.23 Nucleotide sequence identity was calculated by using the Sequence Identity Matrix tool of BioEdit software.23 The phylogenetic analysis was conducted by means of MEGA software, version 6.0,25 the pairwise p-distance assessing the mean nucleotide distances was expressed as crude rate with the corresponding standard deviation.
2.5 Statistical analysis
Statistical analysis was performed by using the Open Source Epidemiologic Statistics for Public Health (OpenEpi; version 3.03).26 The frequency of positive samples was expressed as crude proportion with corresponding 95% confidence interval (95% CI) calculated by Mid-P exact test assuming a normal distribution. Proportions between groups were compared using the Mid-P exact test based on binomial distribution. For continuous variables, the paired t-test was used. P < .05 was considered significant (two-tailed test). The interquartile range (IQR) was computed as difference of first and third quartile of the age distribution.
The start of RSV epidemic season was defined as the 1st week when RSV detection exceed 10% of RSV positivity rate (one gap week was allowed). ILI incidence (per 1000 inhabitants) by week recorded in the considered seasons (from 2014-2015 to 2017-2018) in Lombardy was retrieved from InfluNet.18
3 RESULTS
From 2014-2015 to 2017-2018 influenza season, a total of 1047 ILI cases tested IV-negative out of 2242 samples collected from as many outpatients by the sentinel physicians involved in influenza surveillance network. Information on sex, age, and presence of underling medical conditions of the 1047 ILI outpatients included in this study are detailed in Table 1.
| 2014-2015 | 2015-2016 | 2016-2017 | 2017-2018 | Total | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ILI cases | RSV-positive | ILI cases | RSV-positive | ILI cases | RSV-positive | ILI cases | RSV-positive | ILI cases | RSV-positive | |
| No. of cases; % (95% CI) | 286; 27.3% (24.7%-30.1%) | 34; 11.9% (8.6%-16.1%) | 285; 27.2% (24.6%-30%) | 28; 9.8% (6.8%-13.8%) | 259; 24.7% (22.2%-27.4%) | 40; 15.4% (14.3%-23.7%) | 217; 20.7% (18.4%-23.3%) | 33; 15.2% (11%-20.6%) | 1047; 100% | 135; 12.9% (11%-15.1%) |
| No. of males; % (95% CI) | 156; 54.5% (48.7%-60.2%) | 25; 16% (11.1%-22.6%) | 146; 51.2% (59.6%-70.5%) | 17; 11.6% (7.4%-17.9%) | 125; 48.3% (42.2%-54.3%) | 17; 13.6% (8.7%-20.7%) | 124; 57.1% (50.5%-63.5%) | 18; 14.5% (9.4%-21.8%) | 551; 52.6% (49.6%-55.6%) | 77; 13.9% (11.3%-17.1%) |
| Median age; IQR (range), y | 37.5; 42 (13-55) | 6.5; 45 (3-48) | 34; 43 (8-51) | 37.4; 50.4 (3.9-4.3) | 37.1; 42.4 (13-5.3) | 10.8; 43 (4.4-47.4) | 31.2; 42.4 (9.2-1.7) | 8; 22.1 (3.3-5.6) | 36; 43.4 (10.1-.5) | 8.3; 49 (3-52) |
| No. of cases with underlying medical conditions; % (95% CI) | 55; 19.2% (15%-24.2%) | 5; 10% (3.9%-19.6%) | 56; 19.6% (15.4%-24.6%) | 10; 17.8% (10%-29.8%) | 58; 22.4% (17.9%-28.2%) | 9; 15.5% (8.3%-26.9%) | 55; 25.3% (20%-31.5%) | 7; 12.7% (6.3%-24%) | 224; 21.4% (19%-24.0%) | 31; 13.8% (9.9%-19%) |
| No. of cases by age group; % (95% CI) | ||||||||||
| 0-5 y | 46; 16.1% (12.3%-20.8%) | 16; 34.8% (22.7%-49.2%) | 59; 20.7% (16.4%-25.8%) | 9; 15.2% (8.2%-26.5%) | 38; 14.8% (11%-19.7%) | 13; 34.2% (21.2%-50.1%) | 40; 18.4% (13.8%-24.1%) | 13; 32.5% (20.1%-48%) | 183; 17.5% (15.3%-19.9%) | 51; 27.8% (23.4%-35.3%) |
| 6-15 y | 37; 12.9% (7.5%-14.6%) | 6; 16.2% (7.6%-31.1%) | 42; 14.7% (11.1%-19.3%) | 4; 9.5% (3.7%-22%) | 39; 15% (11.3%-20.1%) | 12; 30.8% (18.6%-46.4%) | 40; 18.4% (13.8%-24.1%) | 8; 20% (10.5%-34.8%) | 158; 15.1% (13%-17.4%) | 30; 18.9% (13.1%-25.1%) |
| 16-45 y | 90; 31.5% (26.4%-37.1%) | 3; 3.3% (1.1%-9.3%) | 97; 34.1% (28.8%-39.7%) | 4; 4.1% (1.6%-10.1%) | 78; 30.1% (25.1%-36.4%) | 5; 6.4% (2.8%-14.1%) | 67; 30.9% (25.1%-37.3%) | 4; 5.9% (2.3%-14.3%) | 332; 31.7% (28.9%-34.6%) | 16; 4.8% (3%-7.7%) |
| 46-65 y | 76; 26.6% (21.8%-32%) | 6; 7.9% (3.7%-16.2%) | 57; 20% (15.8%-25%) | 7; 12.2% (6%-23.2%) | 69; 26.6% (21.9%-32.7%) | 5; 7.2% (3.1%-15.9%) | 47; 21.7% (16.7%-27.6%) | 5; 10.6% (4.6%-22.6%) | 249; 23.8% (21.3%-26.4%) | 23; 9.3% (6.2%-13.5%) |
| >65 y | 37; 12.9% (9.5%-17.3%) | 3; 8.1% (2.7%-21.3%) | 30; 10.5% (7.5%-14.6%) | 4; 13.3% (5.3%-29.7%) | 35; 13.5% (10%-18.4%) | 5; 14.2% (6.2%-29.4%) | 23; 10.6% (7.2%-15.4%) | 3; 13% (4.5%-32.1%) | 125; 11.9% (10.1%-14%) | 15; 12% (7.4%-18.9%) |
- Abbreviations: CI, confidence interval; ILI, influenza-like illness; RSV, respiratory syncytial virus.
Males accounted for 52.6% (n = 551; 95% CI: 49.6%-55.6%) of the study population that had a median age of 36 years with no differences between seasons. A total of 31.7% (n = 332, 95% CI: 28.9%-34.6%) of outpatients belonged to the 16 to 45 years age group, 17.5% (n = 183; 95% CI: 15.3%-19.9%) were children ≤5 years of age, and 11.9% were elderly >65 years (n = 125; 95% CI: 10.1%-14%). 21.4% (224/1047) of ILI cases were individuals with at least one underlying medical condition (Table 1).
A total of 135 (12.9%) samples resulted positive to RSV detection (Table 1). The RSV positivity rate by season ranged between 9.8% (28/285; 95% CI: 6.8%-13.8%) in 2015-2016 and 15.4% (40/259; 95% CI: 14.3%-23.7%) in 2016-2017 (Table 1). As per the general study population, also amongst RSV-positive ILI cases, males (77/135; 57%; 95% CI: 48.6%-65.1%) prevailed females with no significant difference between the two groups (P = .8). RSV-positive ILI cases median age was similar in all considered seasons, with the exception of the 2015-2016 season when it was significantly higher (37.4 years; IQR: 50.4 years; P < .04) (Table 1). Overall, RSV-positive cases were significantly younger than RSV-negative outpatients with a median age of 8.3 years (IQR: 47.8 years) and 37.4 years (IQR: 40.8 years), respectively (P < .001).
RSV was detected in all age groups. In our series of RSV-positive cases, the highest RSV positivity rate was observed in children ≤5 years and in those aged 6 to 15 years, accounting, respectively, for 27.8% (51/183; 95% CI: 23.4%-35.3%) and 18.9% (30/158; 95% CI: 13.1%-25.1%), whereas 12% (15/125; 95% CI: 7.4%-18.9%) of included cases >65 years were RSV-positive (Table 1). Considering only the 33 ILI cases aged less than 1 year, RSV positivity rate was 21.2% (95% CI: 10.6%-37.7%). Among the 224 ILI cases with underlying medical conditions, the RSV positivity rate was 13.8% (31/224; 95% CI: 9.9%-19%), similarly to those without other conditions (104/823; 12.7%; 95% CI: 10.6%-15.2%) (P = .3).
The weekly distribution of RSV-positive and -negative respiratory samples and ILI incidence (per 1000 inhabitants) from 2014-2015 to 2017-2018 season in Lombardy are shown in Figure 1. The median start of RSV epidemic season was in week 48 (ranging from week 46 to week 51), the median peak was in week 8 (ranging from week 6 to week 10); overall, the median width of the RSV epidemics was 15.5 weeks (range: 13-19 weeks).
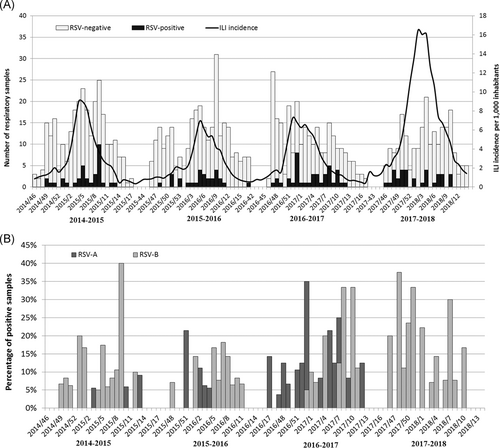
Of the 135 RSV-positive samples, 129 (95.6%) were molecularly characterized. RSV-A and RSV-B were detected in all considered seasons and, overall, RSV-B predominated on RSV-A (63.6% vs 36.4%; P < .001, Table 2 and Figure 1B). In individuals aged 0 to 5 years and in those >65 years, RSV-B prevalence was significantly higher than RSV-A (RSV-A vs RSV-B in 0-5 years: 16.7% vs 83.3%; P = .04; in >65 years 21.4% vs 78.6%; P = .001, Table 2).
| Total of molecularly characterized RSV strains: n = 129; 100% | RSV-A | RSV-B | P-value |
|---|---|---|---|
| No. of total cases; % | 47; 36.4% | 82; 63.6% | <.001 |
| No. of cases by season; % | |||
| 2014-2015 | 6; 18.8% | 26; 81.2% | <.001 |
| 2015-2016 | 12; 46.2% | 14; 53.8% | .5 |
| 2016-2017 | 25; 62.5% | 15; 37.5% | .02 |
| 2017-2018 | 4; 12.9% | 27; 87.1% | <.001 |
| No. of cases by age group; % (95% CI) | |||
| 0-5 y | 1; 16.7% | 5; 83.3% | .04 |
| 6-45 y | 2; 40% | 3; 60% | .6 |
| 46-65 y | 3; 33.3% | 6; 66.7% | .2 |
| >65 y | 3; 21.4% | 11; 78.6% | <.001 |
- Abbreviations: CI, confidence interval; RSV, respiratory syncytial virus.
Both RSV-A and RSV-B were detected in all considered seasons, without a specific trend, as shown in Figure 1B. In particular, RSV-B significantly predominated on RSV-A during both 2014-2015 and 2017-2018 seasons accounting for 81.2% and 87.1% of cases, respectively (P < .001, Table 2), while RSV-A was the subgroup most frequently detected in 2016-2017 season (62.5% vs 37.5%; P = .02, Table 2); in 2015-2016 season, RSV-A and RSV-B co-circulated at similar frequencies (46.2% vs 53.8%; P = .5, Table 2). Co-detections of both RSV-A and RSV-B were not identified in any sample collected from a single patient.
Sequences of the G gene from 27 of 47 (57.4%) RSV-A and 38 of 82 (46.3%) RSV-B positive samples were successfully obtained and aligned with reference sequences of previously published genotypes.24 The majority (23/27; 85.2%) of RSV-A strains belonged to ON1 genotype, and the remaining (4/27; 14.8%) to NA1 genotype. Noteworthy, NA1 strains were identified exclusively in the first two study seasons, while ON1 viruses were detected in all the four considered seasons. Particularly, while the genotypes NA1 and ON1 co-circulated with the same frequency (2/4, 50%) in 2014-2015 season, since 2015-2016 season ON1 genotype (7/9, 77.8%) predominated on NA1 (2/9, 22.2%), then being the only one detected. The mean intragenotypic p-distance of our ON1 strains was 0.020 ± 0.010 for nucleotide sequences. The mean nucleotide identity comparing the ON1 strains detected in this study and the reference strain (isolate ON67-1210A, AN: JN257693) was 98.7% (range: 97.8-99.5%).
All RSV-B strains (n = 38; 100%) belonged to the cluster of BA genotypes. The mean intragenotypic p-distance of our BA strains was 0.021 ± 0.010 for nucleotide sequences. The mean nucleotide identity between BA strains detected in this study and the reference strain (isolate BA/802/99, AN: DQ227363) was 96% (range: 95.1%-97.5%).
4 DISCUSSION
RSV is one of the leading viral causes of acute respiratory infection worldwide affecting human health and public health.1, 5 Most of the recent literature has focused on the impact of RSV in severe infections such as severe acute respiratory infections,27 bronchiolitis,7 and viral pneumonia requiring admission to intensive care units,28 thus hindering a comprehensive evaluation of RSV circulation in general population and its contribution to mild respiratory infections. Besides seasonal IV, several other viral pathogens are involved in clinically undistinguished ILIs, including RSV.5, 29 Well-established influenza surveillance system has been recognized as a valuable tool to monitor the circulation and impact of RSV13 and can be used for RSV surveillance based on the current case definition of ILI.14 Even if a number of European countries have already established a laboratory-based RSV surveillance system, to define the burden, the seasonality and the geographical spread of RSV infection, no RSV surveillance system has been implemented in Italy so far, and only limited information regarding the molecular epidemiology of RSV are available from individual research studies.7, 8, 15-17
This study aimed at investigating the frequency of detection and the molecular epidemiology of RSV in outpatients with ILI in the framework of influenza surveillance during four consecutive winter seasons (from 2014-2015 to 2017-2018). The results obtained from the analysis of 1047 samples collected from ILI cases and tested negative for influenza viruses demonstrated that RSV can significantly contribute to ILI, accounting for nearly 13% of cases, infecting mostly children under 5 years of age, school-aged children and persons over 65 years, and revealing the co-circulation of RSV-A and RSV-B in all considered seasons.
The frequency of RSV detected in this study was similar to that observed in other studies, in which the rate of RSV detection among ILI cases ranged between 7.5% and 11.1%.30, 31 In this study, only IV-negative samples were analyzed, thus not considering the event of RSV/IV coinfection; however, it has been estimated that RSV-IV coinfection account for less than 1.5% of cases in a 7-year study conducted in Austria29 and for 1.2% in another study conducted in Germany.32
The highest incidence of RSV infection is usually reported in children,7, 28, 30, 33, 34 and it is confirmed in our study, where RSV-positivity rate in children younger than 5 years was 27.8%. Moreover, 21% of children under 1 year of age were RSV positive. However, RSV was detected in all considered age groups and it is interesting to note that the median age of RSV-positive individuals was similar in all considered seasons, ranging from 6.5 to 10.8 years, with the exception of 2015-2016 season when the median age of RSV-positive cases raised up to 37.4 years.
It is noteworthy that, in our series, 12% of outpatients over 65 years resulted RSV-positive, thus pointing out the impact of RSV also in the elderly presenting with ILI, as also reported in a systematic review and meta-analysis on RSV impact that has appraised a substantial disease burden of RSV infections among older adults (>65 years).35
A recent study assessing the epidemiology, clinical symptoms, and outcomes related to IV and RSV infections in patients with ILI has demonstrated that RSV has distinct clinical features and outcomes, including a greater impact on individuals with underlining medical conditions.36 Contrary to what resulted from that study, in our series, RSV-positivity rate in ILI cases with or without underlining medical conditions was similar.
RSV circulated throughout the study period (from mid-November to the end of April each season). In the four seasons considered here, the RSV epidemic started at the end of November, with a peak in mid-February, and the median length of the RSV epidemic was nearly 4 months. These findings were similar to those reported by Broberg et al12 who, based on the analysis of data from 15 European countries (not including Italy), have observed that the median start of the RSV epidemic is in week 49, the peak in week 4 and the median length of the epidemic is 13 weeks.12
During RSV epidemic, one subgroup may predominate over the other, or they may circulate concurrently: this variation in RSV-A and RSV-B over time appears to be correlated with the induction of monoclonal antibodies that recognize the RSV primary epitopes, thus indicating immune-driven RSV evolution.4, 34 In our study, RSV-B predominated on RSV-A (63.6% vs 36.4%); this finding is opposed to those observed in recent studies aimed at assessing the distribution of RSV genotypes.7, 9 However, this difference could be a consequence of the study populations considered: in our series all cases were outpatients with ILI while in two previous studies7, 9 included hospitalized young children, in whom over 75% of RSV detected belonged to subgroup A, remarking upon the greater contribution of RSV-A than of RSV-B in severe infection cases.7, 9
During the four consecutive seasons considered in our study, the co-circulation of RSV-A and RSV-B was observed, as previously described by others.7, 34 In details, in 2014-2015 season, RSV-B predominated on RSV-A, followed by a season characterized by a similar circulation of both subgroups, then, in the next season (2016-2017) RSV-A predominated on RSV-B, which was predominant also in the 2017-2018 season.
Multiple RSV genotypes can circulate simultaneously in a community7, 15, 34; in this study, the majority (85%) of RSV-A belonged to genotype ON1, while the remaining were NA1 (15%). In the 2014-2015 season, NA1 and ON1 genotypes circulated at the same extent, and in the 2015-2016 season the ON1 predominated on the NA1 that was completely replaced by the ON1 genotype from 2015-2016 season ahead. Genotype ON1—characterized by a 72-nt duplication in the second hypervariable region of the G gene—was first described in Canada in 201037 and, as observed in other studies,7-9 it has rapidly spread, replacing the ancestor NA1. All RSV-B strains identified in our study belonged to genotypes BA cluster; these genotypes—with a 60-nt duplication in the second hypervariable region of the G gene—were first described in Buenos Aires in 1999,38 and then they become the predominant RSV-B genotypes circulating in Europe in recent years.7-9, 32
As declared by the World Health Organization RSV surveillance group—that leverages the capacities of the Global Influenza Surveillance and Response System—countries with existing influenza surveillance systems are able to build on RSV surveillance on top of their influenza surveillance program.39 However, the lack of a uniform case definition for RSV still poses a challenge to characterize the epidemiology, clinical features, and disease burden of RSV, thus complicating the interpretation of surveillance results.39 In our study, the standard case definition of ILI proposed by the European Centre for Disease Prevention and Control,14 which includes an abrupt onset of fever (>38°C) or feverishness, was used, as also suggested by others12, 13; however, it has been shown that including fever in a case definition for RSV may lower the sensitivity to detect cases.13, 39 This means that, in interpreting our results, the RSV burden should have been underestimated. In addition, another limitation of our study is that sampling time was restricted to influenza season, thus the prevalence of RSV was estimated only in the period spanning from November to April.
In conclusion, investigating the epidemiological and molecular characteristics of RSV in ILI cases can increase the current knowledge of RSV epidemiology and impact that are essential in the perspective to set up specific vaccination strategies.
ACKNOWLEDGMENTS
The authors would like to thank the general practitioners and pediatricians involved in the Italian Influenza Surveillance Network for Lombardy region.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.