Reverse transcriptase droplet digital PCR vs reverse transcriptase quantitative real-time PCR for serum HBV RNA quantification
Abstract
Serum hepatitis B virus (HBV) RNA is a novel marker reflecting the activity of covalently closed circular DNA. However, the methodology for detecting HBV RNA has been a technical challenge. In this study, the performance of reverse transcription droplet digital polymerase chain reaction (RT-ddPCR) for quantifying HBV RNA was compared with that of reverse transcription quantitative real-time PCR (RT-qPCR) in serum samples collected from treatment-naïve patients with different phases of chronic hepatitis B (CHB). A total of 417 serum samples, including 136 HBeAg-positive CHB and 281 HBeAg-negative CHB were examined. HBV RNA levels measured by RT-ddPCR and RT-qPCR showed a high degree of linearity and quantitative correlation. The limit of detections of RT-ddPCR and RT-qPCR assays were 102 and 103 copies/mL, respectively. Our results also demonstrated that RT-ddPCR was superior to RT-qPCR in terms of its consistency for quantifying HBV RNA across all concentrations. In the HBeAg-positive group, serum HBV RNA levels based on RT-ddPCR were moderately correlated with HBV DNA (r = 0.591, P < .001) and HBsAg (r = 0.502, P < .001). Among patients with HBeAg-negative CHB, serum HBV RNA levels were moderately correlated with HBV DNA (r = 0.603, P < .001) but had weak correlation with HBsAg (r = 0.203, P = .001). In summary, RT-ddPCR could enhance the sensitivity of serum HBV RNA detection, particularly among the HBeAg-negative group with low viral loads. Thus, RT-ddPCR could serve as an optimal method for HBV RNA quantification in clinical practice.
Highlights
-
RT-ddPCR vs RT-qPCR for serum HBV RNA quantification.
Abbreviations
-
- ALT
-
- alanine aminotransferase
-
- AST
-
- aspartate aminotransferase
-
- CHB
-
- chronic hepatitis B
-
- HBeAg
-
- hepatitis B e antigen
-
- HBsAg
-
- hepatitis B surface antigen
-
- HBV
-
- hepatitis B virus
-
- RT-ddPCR
-
- reverse transcription droplet digital PCR
-
- RT-qPCR
-
- reverse transcription quantitative real-time PCR
1 INTRODUCTION
Hepatitis B virus (HBV) is a major etiological cause of chronic hepatitis, which may progress to cirrhosis and the development of hepatocellular carcinoma (HCC).1 HBV is a partially double-stranded DNA virus that replicates via the organization of covalently closed circular DNA (cccDNA). In infected hepatocytes, cccDNA acts as the template for pregenomic (pg)RNA transcription and viral protein production.2 Current evidence has indicated that the persistence of intrahepatic cccDNA is responsible for disease chronicity and represents a major barrier for HBV eradication.3 Thus, quantitative evaluation of intrahepatic cccDNA and its transcriptional activity is considered to be essential for the management of patients with chronic hepatitis B (CHB).4 Despite its essential role in viral replication, the measurement of cccDNA in clinical practice is restricted to the requirement for liver biopsy and lack of available commercial assays.
Recently, serum HBV RNA quantification has emerged as a novel biomarker of HBV infection.5, 6 It has been demonstrated that circulating HBV RNA is well correlated with the transcriptional activity of intrahepatic cccDNA7, 8 and displays unique patterns in the natural history of chronic HBV infection.8 In untreated cases, circulating HBV RNA levels have a good correlation with levels of HBV DNA. In patients undergoing nucleos(t)ide analogue (NA) therapy, although HBV DNA is suppressed, HBV RNA is unaffected. Therefore, HBV RNA appears to be a reliable biomarker representing cccDNA activity.6 The detectable serum HBV RNA level is also correlated with risk of HBV rebound after withdrawal of NAs in patients with CHB.6 In addition, baseline and kinetics of HBV RNA levels can predict HBeAg seroconversion with higher accuracy than those of HBV DNA levels.9 Therefore, serum HBV RNA might be a potential biomarker for disease monitoring, predicting treatment response and prognostic outcome of patients with CHB.
Presently, the principal technique for measuring serum HBV RNA is reverse transcriptase quantitative polymerase chain reaction (RT-qPCR).5, 6, 9 However, RT-qPCR needs more than 103 copies of HBV RNA to produce a good quantitative signal.6 Thus, this limitation might compromise the diagnostic performance of this method, particularly among samples with very low HBV RNA levels such as HBeAg-negative CHB. Droplet digital PCR (ddPCR) is a recent approach based on water–oil emulsion droplet technology that has been increasingly used for DNA/RNA quantification.10, 11 The ddPCR is created on the combination of traditional PCR and fluorescent-probe-based detection techniques to allow highly sensitive absolute quantification of nucleic acids without the requirement for standard curves. Following this method, a sample is partitioned into thousands of droplets, then PCR amplification occurs within each droplet. As a result, the ddPCR technology can improve the sensitivity and enable the detection of low concentrations of DNA/RNA samples compared with qPCR technique.12 Thus, this technology may represent an ideal assay for measuring serum HBV RNA levels in clinical practice. Thus, the aim of this study was to evaluate the potential role of RT-ddPCR for HBV RNA quantification in patients with CHB and compare its performance to that of RT-qPCR assay.
2 MATERIALS AND METHODS
2.1 Clinical samples
A total of 417 serum samples from Thai patients with chronic HBV infection were included in this study. The diagnosis of chronic HBV infection was confirmed by the presence of serum hepatitis B s antigen (HBsAg) in the previous 6 months. All patients were naïve to antiviral treatment and were recruited from King Chulalongkorn Memorial Hospital in Bangkok during their first visit. Patients were classified according to serum hepatitis B e antigen (HBeAg) status, which was either HBeAg-positive CHB (n = 136) or HBeAg-negative CHB (n = 281). Exclusion criteria included patients who were seropositive for hepatitis C virus or human immunodeficiency virus, and those who had clinical evidence of HCC diagnosed by imaging studies. Written informed consents were obtained from patients. This study was approved by the Institutional Review Board of the Faculty of Medicine of Chulalongkorn University (IRB number 016/61).
2.2 RNA extraction and complementary DNA synthesis
Total RNA was extracted from 200 µL of serum samples as previously described.13 Briefly, cells were lysed with denaturing solution (4M guanidine thiocyanate, 25 mM sodium citrate [pH 7.0], 0.5% (wt/vol) N-laurosylsarcosine, and 0.1M 2-mercaptoethanol) followed by phenol/chloroform extraction and isopropanol precipitation. The RNA pellet was dissolved in 20 uL of diethylpyrocarbonate-treated water and treated with DNAse (DNase I, RNase-free; Thermo Fisher Scientific) according to the manufacturer's instructions. Then, 4 uL of RNA was used as input in a reverse transcription reaction in a 20 uL total reaction volume by using ImProm-II™ Reverse Transcription System (Promega) and HBV-specific RT primer as previously described.6 The sequence of HBV-specific RT primer is 5′ATTCTCAGACCGTAGCACACGACACCGAGATTGAGATCTTCTGCGAC-3′ in which the random sequence ATTCTCAGACCGTAGCACACGACAC was anchored at the 5′ end of the HBV-specific sequence CGAGATTGAGATCTTCTGCGAC. The same complementary DNA (cDNA) preparations were used for both ddPCR and qPCR to ensure consistency.
2.3 Construction of plasmids used as positive controls
Standard plasmids containing specific HBV cDNA target fragments were used as template for the optimization of the assays in serum HBV RNA quantification. Plasmids were constructed by inserting the DNA fragment encoding the HBV core region into pGEM-T Easy Vector (Promega, Madison, WI) using the TA-cloning strategy. The resulting plasmid constructs were confirmed by PCR and DNA sequencing.
2.4 TaqMan-based quantitative real-time polymerase chain reaction
The levels of HBV RNA were determined as previously described.6 The serum HBV RNA was quantified by qPCR in ViiA 7 Real-Time PCR System (Applied Biosystems, Foster City, CA) with TaqMan probe method. The sequences of the primers and probe were forward primer 5′-AYAGACCATCAAATGCCC-3′, reverse primer (Anchored sequence) 5′ATTCTCAGACCGTAGCACACGACAC-3′, and probe 5′ FAM-CTTATCAACACTTCCGGARACTACTGTTGTTA GAC-BHQ1-3′. The 10 uL qPCR reaction mixture contained 5 uL of 2X Master Mix (QPCR Probe Master Mix LRox; Biotechrabbit, Germany), 0.4 uL of 10 uM forward primer, 0.4 uL of 10 uM reverse primer, 0.4 uL of 5 uM TaqMan probe, 2 uL cDNA template, and 1.3 uL water. Cycling parameters consisted of denaturation at 95°C for 3 minutes, followed by 40 cycles at 95°C for 15 seconds, and 60°C for 30 seconds. The level of HBV cDNA was quantified by comparing the signals to a standard curve using Applied Biosystems QuantStudio Real-Time PCR system software. To evaluate HBV DNA carryover, qPCR was performed in parallel on RNA samples without reverse transcription (no-RT controls). For samples with high HBV RNA levels above the upper limit of quantification, the appropriate dilutions of cDNA template were performed.
2.5 Droplet digital PCR
Serum HBV-RNA was quantified using ddPCR platform (QX200; Bio-Rad). A total of 20 uL reaction mixture included 10 uL of 2X ddPCR Supermix (Bio-Rad), 1.8 uL of 10 nM primers, 1.0 uL of TaqMan Probe, 2 uL of the cDNA template, and 3.4 uL of water. The sequences of primer and probe were the same as in the qPCR system. For each ddPCR reaction mixture, 70 uL of droplet generation oil was added to the DG8 cartridge and the droplets were produced by a droplet generator of the QX200 Droplet Digital PCR system (Bio-Rad). The droplets contained the PCR reaction mixture and droplet generation oil, which were transferred to a 96-well PCR plate for amplification using the T100 Thermal Cycler (Bio-Rad). The experiments were performed using the following protocol: one cycle at 95°C for 10 minutes, 40 cycles at 94°C for 30 seconds and 55°C for 1 minute and one cycle at 98°C for 10 minutes. Subsequently, a droplet reader was used to calculate the number of both positive and negative droplet events from each PCR reaction mixture. A PCR reaction mixture with no DNA template was used as a reference control for potential PCR contamination. The concentration of cDNA was calculated by QuantaSoft analysis software (Bio-Rad). The ddPCR data and the concentration of positive droplets were determined by calculating the ratio of the positive droplets over the total droplets combined with Poisson distribution. For samples with HBV RNA levels higher than 106 copies, the appropriate dilutions of cDNA template were performed. The process of no-RT controls for this assay is shown in Figure S1.
2.6 Serological and virological assays
Qualitative measurements of conventional serum HBV markers including HBsAg, HBeAg, anti-HBe, and anti-HBs were determined by commercially available enzyme-linked immunosorbent assays (Abbott Laboratories, Chicago, IL). Serum HBsAg quantification was assessed by Elecsys HBsAg II Quant reagent kits (Roche Diagnostics, Indianapolis, IN) and serum HBV DNA level was quantified by Abbott Real Time HBV assay (Abbott Laboratories).
2.7 Data analysis and statistics
Statistical analysis was performed using SPSS statistics software 22.0 for Windows (SPSS Inc, Chicago, IL) and GraphPad Prism v5.0 (GraphPad Software, San Diego, CA). Comparisons between groups were assessed by the χ2 or Fisher's exact test for categorical variables and by the Mann–Whitney U-test or Student t-test for quantitative variables as appropriate. Correlation between parameters was examined by the Spearman rank correlation test. P < .05 was considered statistically significant.
3 RESULTS
3.1 Patient characteristics
Baseline demographic data of 417 patients according to HBeAg status are shown in Table 1. Sex distribution between the HBeAg-positive and HBeAg-negative CHB groups was similar, although the HBeAg-positive patients were significantly younger. The latter also had higher levels of serum aspartate aminotransferase (AST), alanine aminotransferase (ALT), viral markers (HBV DNA and HBsAg), and liver stiffness measurement (LSM) by FibroScan compared to the HBeAg-negative group.
| HBeAg-positive CHB | HBeAg-negative CHB | ||
|---|---|---|---|
| Characteristics | (n = 136) | (n = 281) | P-value |
| Age, y | 35.5 ± 10.4 | 48.6 ± 12.1 | <.001 |
| Sex (male) | 76 (55.9%) | 147 (52.3%) | .530 |
| AST (U/L) | 50.3 ± 37.4 | 26.5 ± 14.9 | <.001 |
| ALT (U/L) | 72.7 ± 65.0 | 36.3 ± 32.6 | <.001 |
| Log10 HBV DNA (IU/mL) | 7.9 ± 1.2 | 4.0 ± 1.7 | <.001 |
| Log10 HBsAg (IU/mL) | 4.2 ± 0.7 | 2.7 ± 1.3 | <.001 |
| Liver stiffness (kPa) | 6.9 ± 4.0 | 6.0 ± 2.6 | .033 |
- Note: Data described as means ± SD or n (%)
- Abbreviations: ALT, alanine aminotransferase; AST, aspartate aminotransferase; CHB, chronic hepatitis B; HBeAg, hepatitis B e antigen; HBsAg, hepatitis B s antigen; HBV, hepatitis B virus.
3.2 Optimization of the RT-ddPCR assay
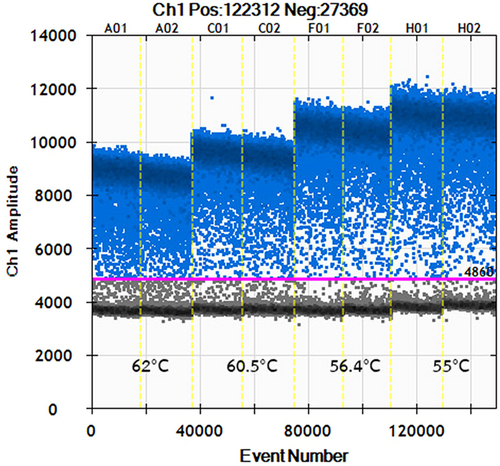
To determine the optimal annealing temperature for RT-ddPCR assay, we tested different annealing temperatures at 55, 56.4, 60.5, and 62.0°C. The annealing temperature at 55°C yielded the largest difference in fluorescence between negative and positive droplets (Figure 1). Lower annealing temperatures resulted in nonspecific amplifications, which might overestimate the quantitation of HBV RNA. Therefore, an optimized annealing temperature of 55°C was chosen for the subsequent RT-ddPCR tests.
3.3 Comparison of analytical sensitivity, linearity, and dynamic range between RT-ddPCR and RT-qPCR assays
Ten-fold serial dilution of plasmid control was subjected to testing for analytical sensitivity, linearity, and dynamic range of the two assays. The RT-qPCR assay exhibited good linearity (R2 = 0.998) with dynamic range of detection from 103 to 108 copies/mL (Figure 2A). The slope was −3.36, equivalent to a RT-PCR efficiency of 98.3%. According to the standard curves, the detection limit of RT-qPCR assays was determined as 103 copies/mL. Quantitative linearity of RT-ddPCR assay was also assessed by positive plasmid DNA. The log10-transformed copy numbers concentration measured by RT-ddPCR were plotted against the corresponding log10-transformed predicted values of serially diluted plasmid DNA and fitted with a linear regression mode. The measurements of RT-ddPCR assay exhibited good linearity (R2 = 0.995, P < .001) between the target input amounts and measured values in a dynamic range of 106 to 102 copies/mL (Figure 2B). However, when the input of DNA template was higher than 106 copies, the RT-ddPCR droplets became completely saturated. In this study, the sensitivity of the RT-ddPCR assay for plasmid DNA was down to 102 copies/mL, which was more sensitive than RT-qPCR. Importantly, no nonspecific amplifications were observed in the negative controls for both assays.

3.4 Evaluation of repeatability between RT-ddPCR and RT-qPCR assays
To evaluate and compare the reproducibility of the RT-ddPCR and RT-qPCR assays, we serially diluted cDNA extracted from serum samples. From the starting HBV RNA quantity of approximately 3338 copies/mL by RT-ddPCR, we generated a moderate concentration (approximately 668 copies/mL), low concentration (approximately 134 copies/mL), and very low concentration (approximately 27 copies/mL) dilutions of cDNA. Then, the diluted cDNA was subjected to both assays. Each assay was performed five times in the same run. The coefficient of variation (CV) was then calculated (Table 2).
| RT-ddPCR | RT-qPCR | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Samplea (copies/mL) | pos/total | Copies/mL | Mean ± SD | CV (%) | pos/total | Copies/mL | Mean ± SD | CV (%) | P-values |
| High concentration (3338) | 5/5 | 2963; 3300; 3375; 3675; 3338 | 3330 ± 254 | 7.6 | 5/5 | 3937; 4340; 5118; 3075; 3819 | 4057.9 ± 748.6 | 18.4 | .073 |
| Moderate concentration (668) | 5/5 | 750; 675; 551; 863; 713 | 710 ± 113 | 15.9 | 5/5 | 739; 957; 1214; 927; 908 | 948.9 ± 170.4 | 18.0 | <.05 |
| Low concentration (134) | 5/5 | 75; 26; 75; 49; 49 | 55 ± 21 | 37.7 | 5/5 | 112; 116; 119; 255; 91 | 138.6 ± 66.0 | 47.7 | <.05 |
| Very low concentration (27) | 4/5 | 0; 26; 23; 71; 26 | 29 ± 26 | 88.5 | 3/5 | 0; 0; 36; 29; 35 | 20.1 ± 18.5 | 92.1 | .539 |
| Negative control | 0/5 | 0; 0; 0; 0; 0 | 0 | N/A | 0/5 | 0; 0; 0; 0; 0 | 0 | N/A | N/A |
- Abbreviations: CV, coefficient of variation; HBV, hepatitis B virus; pos/total, positive/total; RT-ddPCR, reverse transcription droplet digital polymerase chain reaction; RT-qPCR, reverse transcription quantitative real-time polymerase chain reaction
- a The serum sample was obtained from an HBV-infected patient and was quantified as approximately 3338 copies/reaction by RT-ddPCR. The cDNA was diluted to moderate concentration (approximately 668 copies/mL), low concentration (approximately 134 copies/mL), and very low concentration (approximately 27 copies/mL).
The RT-ddPCR yielded 100% positive results for low concentration samples and 80% positive results for very low concentration samples. The corresponding figures for RT-qPCR were 100% and 60%, respectively. At all concentrations, HBV RNA levels detected by RT-ddPCR were less variable than those of RT-qPCR. No nonspecific amplifications of the negative control were observed in either assay.
3.5 Evaluation of clinical samples
All 417 serum samples from patients with CHB were tested by RT-ddPCR and RT-qPCR assays. Serum HBV RNA was detected in 170 of 417 (40.8%) of patients by RT-qPCR, including 132 of 136 (97%) in the HBeAg-positive group and 38 of 281 (13%) in the HBeAg-negative group. In contrast, RT-ddPCR assay could detect serum HBV RNA in 279 of 417 (67%) of patients, which included 136 of 136 (100%) of the HBeAg-positive group and 143 of 281 (51%) of the HBeAg-negative group (Table 3). Thus, our data showed that RT-ddPCR assay was more sensitive than RT-qPCR technique in detecting serum HBV RNA, particularly in patients with HBeAg-negative CHB.
| RT-ddPCR | RT-qPCR | ||||||
|---|---|---|---|---|---|---|---|
| Log10copies/mL | Log10copies/mL | ||||||
| Patients | pos/total (%) | Mean ± SD | Range | pos/total (%) | Mean ± SD | Range | P-value |
| HBeAg-positive CHB | 136/136 (100.0) | 6.57 ± 1.46 | 2.72-9.94 | 132/136 (97.1) | 6.60 ± 1.29 | 3.31-9.48 | .083 |
| HBeAg-negative CHB | 143/281 (50.9) | 3.71 ± 1.10 | 2.65-7.74 | 38/281 (13.5) | 4.39 ± 1.03 | 3.10-7.43 | <.001 |
| Total | 279/417 (66.9) | 5.10 ± 1.93 | 2.65-9.94 | 170/417 (40.8) | 6.10 ± 1.54 | 3.10-9.48 | <.001 |
- Abbreviations: CHB, chronic hepatitis B; HBeAg, hepatitis B e antigen; HBV, hepatitis B virus; RT-ddPCR, reverse transcription droplet digital polymerase chain reaction; RT-qPCR, reverse transcription quantitative real-time polymerase chain reaction.
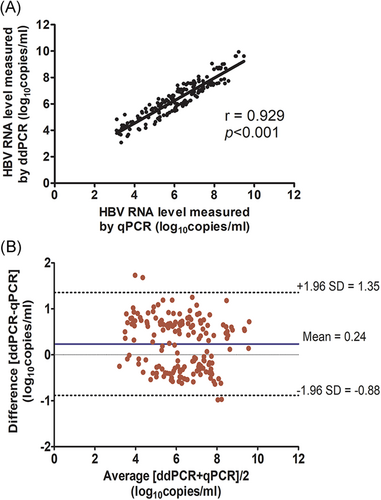
Among 170 samples that were positive for both tests, their correlation curve was generated (Figure 3A). The Pearson r coefficient was 0.929 (P < .001). In addition, Bland–Altman plots comparing the mean quantitative values for RT-ddPCR and RT-qPCR assays demonstrated a close agreement between the two methods (means ± SD, 0.24 ± 0.57 log10copies/mL) (Figure 3B).
3.6 Correlation of hepatitis B virus RNA levels with clinical and viral parameters
Overall, HBV RNA levels based on RT-ddPCR assay were moderately correlated with serum AST (r = 0.476, P < .001), ALT (r = 0.443, P < .001), HBV DNA (r = 0.838, P < .001), and serum HBsAg (r = 0.597, P < .001). In addition, HBV RNA levels correlated weakly with LSM (r = 0.229, P < .001), but exhibited a negative correlation with age of the patients (r = −0.498, P < .001).
Stratification based on the HBeAg status showed that the mean HBV RNA level in the HBeAg-positive group was significantly higher than in the HBeAg-negative groups (6.6 ± 1.5 vs 1.9 ± 2.0 log10copies/mL, P < .001). In the HBeAg-positive group, serum HBV RNA levels were negatively correlated with age (r = −0.261, P = .002), but positively correlated with serum HBV DNA (r = 0.591, P < .001) and serum HBsAg (r = 0.502, P < .001). Serum HBV RNA did not correlate with AST (r = 0.042, P = .657), ALT (r = 0.056, P = .522), and LSM (r = 0.165, P = .100). Among patients with HBeAg-negative CHB, serum HBV RNA levels correlated with age (r = −0.230, P < .001), AST (r = 0.268, P < .001), ALT (r = 0.403, P < .001), LSM (r = 0.169, P = .007), HBV DNA (r = 0.603, P < .001), and HBsAg (r = 0.203, P = .001).
4 DISCUSSION
Earlier studies have demonstrated that serum HBV RNA quantification may serve as a novel surrogate marker for intrahepatic cccDNA transcriptional activity,8, 14 and could represent a useful marker for assessment of antiviral efficacy.5, 9, 15 In recent years, various RT-qPCR-based quantitative methods for detecting serum HBV RNA have been developed. In this study, we directly compared the performance of RT-ddPCR quantification with RT-qPCR assay in clinical samples of patients regarding HBeAg status in the natural history of HBV infection. We demonstrated that both methods displayed a high degree of linearity and excellent quantitative correlation in clinical samples of patients with CHB. However, RT-ddPCR assay had better sensitivity and reproducibility than RT-qPCR platform, particularly in patients with HBeAg-negative CHB. Specifically, RT-ddPCR assay could allow the quantitation of serum samples with HBV RNA levels as low as 102 copies/mL. In contrast, the cut-off level of RT-qPCR assay was approximately 103 copies/mL. As a result, when applied to the samples of patients with HBeAg-negative CHB, a considerably higher positive result was obtained by RT-ddPCR method compared with RT-qPCR technique (51% vs 13%, respectively). Of note, the lower limit of detection for HBV RNA by RT-qPCR in our study was comparable to a recent report using a rapid amplification of cDNA ends qPCR technique.16 Our data also support previous studies demonstrating that RT-ddPCR assay is more sensitive than RT-qPCR method in clinical settings.17-19
The lower cut-off level of RT-ddPCR assay developed here that was based on a TaqMan hydrolysis probe method was in line with a recent study by Wang et al.20 In that study, a DNA binding dye (EvaGreen)-based ddPCR assay could determine HBV RNA levels in NA-treated patients as low as 200 copies/mL. Collectively, our findings demonstrated that RT-ddPCR offered an improved HBV RNA quantification, which is particularly appropriate for detecting specimens with very low copy numbers. This benefit could potentially facilitate decision-making in terms of accurate clinical assessment in patients undergoing antiviral therapy. In this respect, recent data have shown that HBV RNA measurement could represent a good predictor for discontinuation of NA therapy in patients with CHB.21 In a multicenter prospective cohort study, for instance, non-cirrhotic patients who were double negative for HBV-DNA and HBV-RNA at the end of NA treatment had significantly lower risk of viral relapse in long-term follow-up compared with those who had positive serum HBV-DNA or HBV-RNA levels.22 As a result, a more sensitive and reliable method for HBV RNA assay may assist decisions to stop NA therapy.
Our data confirmed previous reports that there were diverse patterns of serum HBV RNA levels during the natural history of HBV infection.8, 23 In this study, mean HBV RNA level significantly differed by HBeAg status as levels were obviously higher in the HBeAg-positive group compared with those in the HBeAg-negative group. These findings likely indicate a higher degree of transcriptional activity of cccDNA in patients with HBeAg-positive CHB as compared to patients with HBeAg-negative CHB. In addition, we compared the correlation of serum HBV RNA levels with other HBV markers, including HBV DNA and HBsAg quantification. In the HBeAg-positive group, serum HBV RNA levels were moderately correlated with serum HBV DNA (r = 0.591, P < .001) and serum HBsAg quantification (r = 0.502, P < .001). In patients with HBeAg-negative CHB, however, serum HBV RNA displayed a moderate correlation with serum HBV DNA (r = 0.603, P < .001) but showed a weak correlation with serum HBsAg (r = 0.203, P = .001). Such correlation between serum HBV RNA and HBsAg levels according to HBeAg status was similar to recent reports.16, 23 The weak correlation between these two markers in HBeAg-negative CHB might reflect high complexity of HBsAg expression in this subgroup of patients as the production of this viral protein is not derived only from the transcriptional activity of cccDNA, but also from integrated viral sequences the host genome.24
This study had some limitations including being a cross-sectional study and the samples obtained from Thai individuals were mainly genotype B or C. Previous reports revealed that HBV genotypes might have an influence on serum HBV RNA levels,16 thus the results obtained from our study might not be directly applicable to other HBV genotypes. In summary, our data demonstrated that RT-ddPCR method could increase the rate of HBV RNA detection from samples with low viral loads, thereby improving the diagnosis and management of patients with CHB. As growing evidence supports the usefulness of serum HBV RNA, the development of reliable and reproducible quantification of this novel marker, as well as its validation in clinical practice, deserve further exploration.
ACKNOWLEDGMENTS
This study was funded by The Thailand Research Fund (TRF) Senior Research Scholar (RTA6280004), the Grant for Chula Research Scholar (CU-GRS-61-07-30-02), the 90th Anniversary of Chulalongkorn University Fund (Ratchadaphiseksomphot Endowment Fund), and the Second Century Fund (C2F). The study was also supported by Center of Excellence in Hepatitis and Liver Cancer, The Center of Excellence in Clinical Virology of Chulalongkorn University, and the Research Chair Grant from NSTDA (P-15-50004). The authors thank Sompong Vongpunsawad (Center of Excellence in Clinical Virology, Chulalongkorn University) for reviewing this manuscript.