Human pegivirus-1 infection in kidney transplant recipients: A single-center experience
Abstract
Kidney transplantation is the treatment of choice for patients with end-stage renal disease. In the posttransplant period, the induced immunosuppression leads to an increased risk of developing infectious diseases, a leading cause of death after kidney transplantation. Human pegivirus-1 (HPgV-1) is considered a nonpathogenic human virus and is highly frequent in individuals parenterally exposed, however, its impact on kidney transplantation outcome is poorly understood. Given the scarcity of epidemiological data for this infection on organ recipients in Brazil, we conducted a study in a single center for kidney transplantation in Rio de Janeiro, aiming to determine HPgV-1 prevalence and genotypic distribution. Serum samples from 61 renal recipients, followed up for the first year after transplantation, were evaluated for viral RNA and genotypes were determined by sequencing of the 5′-untranslated region. HPgV-1 RNA was detected in 36.1% (22/61) of patients. Genotype 2 was the most commonly found (80.9%), followed by genotypes 3 (9.5%), 1, and 5, in 4.8% each. Statistical comparisons did not reveal any significant impact of HPgV-1 in patient outcome. Further epidemiologic studies are needed to understand if immunosuppression may interfere in HPgV-1 persistence rates and if viremia might impact graft dysfunction rates in kidney recipients.
Highlights
-
High prevalence of HPgV-1-RNA (36.1%) was found in kidney transplant recipients.
-
No significant impact of HPgV-1 infection in patient outcome was observed.
-
High HPgV-1 persistence rates were observed among kidney recipients patients.
-
Genotype 2 was predominantly found, followed by genotypes 3, 1 and 5.
1 INTRODUCTION
Chronic kidney disease (CKD) is a global public health issue that affects approximately 120,000 people in Brazil.1 Kidney transplantation is the treatment of choice for the majority of patients with CKD conferring better survival and long-term quality of life when compared to patients undergoing dialysis.2 Several factors have contributed for better prognosis for patients undergoing kidney transplantation, as improvement in surgical techniques and immunosuppressive therapy. Even though modern immunosuppressive therapy has dramatically reduced the incidence of acute rejection and improved graft survival, it is also associated with various posttransplant complications.3 In developed countries, cardiovascular events are the major cause of death posttransplant, but in Brazil, the major cause of death is infection, not only during the first year but during any time after kidney transplantation.4
Human pegivirus-1 (HPgV-1, previously denominated GB virus C/hepatitis G virus) is a single-stranded RNA virus classified into genus pegivirus within the Flaviviridae family. The genomic organization of pegivirus features a 9.4-kb RNA genome, 5′- and 3′-untranslated regions (UTR), and an open-reading frame encoding for a polyprotein subsequently cleaved by proteases to produce functional and structural proteins.5 This genus was formerly divided into two species (pegivirus A and pegivirus B), but recently, its classification was revised and new species were proposed. HPgV-1 is currently classified into pegivirus C and the newly discovered HPgV-2 is classified into pegivirus H species.6
On the basis of phylogenetic analyses of 5′-UTR, E2, or complete genomic sequences, at least six genotypes of the virus have been described. Geographically, these genotypes show distinct distribution patterns. In general, genotype 1 is predominant in Africa, genotype 2 is found in Europe and America. Genotype 3 is the most common in Asia including Japan and China. Genotype 4 is predominant in Southeast Asia, and genotype 5 is only seen in South Africa and genotype 6 in Indonesia. Recently, genotype 7 was proposed with its sequences discovered in China.7, 8
HPgV-1 is transmitted through parenteral, sexual, and perinatal routes. Worldwide, ~750 million people are actively infected and HPgV-1 viremia in blood donors from developed countries varies from 1% to 5%, whereas is higher in the same populational group in developing countries (up to 20%).9, 10 Due to shared modes of transmission, HPgV-1 viremia is highly prevalent in human immunodeficiency virus (HIV)- and hepatitis C virus (HCV)-infected or parenterally exposed individuals, as hemodialysis patients or organ transplantation receptors.11 HPgV-1 commonly causes persistent infection and to date it has not been shown to cause any human disease. As a commensal virus, any change in immunological status, as immunosuppression received after an organ transplantation, may shift the condition between innocuous viral resident and disease-causing pathogen. There is already evidence that complications experienced by haematopoietic stem cell transplant recipients may be modulated by the presence of commensal viruses.12 The aim of this study is to determine the prevalence and genotypic distribution of HPgV-1, a nonpathogenic human virus, among kidney transplant recipients as well as evaluate its role in patient outcome.
2 MATERIALS AND METHODS
2.1 Study population
This study was carried out in one center for organ transplantation in Rio de Janeiro and included 61 patients who underwent kidney transplantation between April and August 2015. Blood samples were collected with 3-month intervals, up to 1-year period, as part of their follow up after renal transplantation. In total, 257 blood samples were submitted to molecular analyses, representing a mean of approximately four samples per patient. In two cases, only one sample was available due to the occurrence of allograft rejection before the second collection point. Data were collected from the patients’ medical records. Demographic data included age (donor and receptor), sex, cause of CKD, and immunosuppressive therapy adopted. Patients were also screened for hepatitis B virus (HBV), HCV, and HIV serological markers before transplant. This study was conducted in accordance with the Declaration of Helsinki and the protocol was approved by the Research Ethics Committee of the Clementino Fraga Filho University Hospital (approval number 1.211.280/2015).
2.2 Nucleic acid extraction and detection of HPgV-1 RNA
Viral RNA was extracted from 200 µL plasma samples using High Pure Viral Nucleic Acid Kit (Roche Diagnostics GmbH, Mannheim, Germany) according to the manufacturer's instructions. Detection of viral RNA was performed by nested reverse transcription polymerase chain reaction (RT-PCR) using SuperScript III One-Step RT-PCR System with Platinum Taq DNA Polymerase Kit (Invitrogen, Carlsbad, CA) with specific primers for partial 5′-UTR amplification. Nucleotide positions are numbered according to reference sequence U44402, as described by Jarvis et al.13 Briefly, 5 µL from extracted RNA were added to 20 µL of a reaction mixture containing 2X reaction mix, 10 µM of outer sense primer 108 (5′-AGGTGGTGGATGGGTGAT-3′), 10 µM of outer antisense primer 531 (5′-TGCCACCCGCCCTCACCCGAA-3′), 1 µL SuperScript III RT/Platinum Taq Mix, and 5.5 µL of autoclaved distilled water. One-step RT-PCR amplification consisted of a preincubation at 50°C for 20 minutes and 94°C for 2 minutes, followed by 40 cycles with the following incubation times and temperatures: 94°C 30 seconds, 50°C 30 seconds, and 68°C 30 seconds. For the second round, 5 µL of first round product were added to 45 µL of a reaction mixture containing: 1X buffer, 0.2 µM dNTP mixture, 1.5 mM MgCl2, 10 µM of inner sense primer 134 (5′-TGGTAGGTCGTAAATCCCGGT-3′), 10 µM of inner antisense primer 476 (5′-GGAGCTGGGTGGCCCCATGCAT-3′), 1U Taq DNA Polymerase (Invitrogen), and autoclaved distilled water. Amplification of a 344-bp fragment was performed under following conditions: 94°C 2 minutes, 40 cycles at 94°C 30 seconds, 60°C 30 seconds, 72°C 30 seconds, and a final extension at 72°C 5 minutes.14 Ten microliters of amplification product were loaded on 2% agarose gels, electrophoresed, stained with ethidium bromide, and visualized under UV light.
2.3 Sequencing and genotyping
The 344-bp PCR product was purified with the High Pure PCR Product Purification Kit (Roche Diagnostics) and directly sequenced using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City) according to the manufacturer's protocol. The retrieved electropherogram from both strands were analyzed and edited using MEGA 7 software15 to generate consensus sequences. HPgV-1 sequences obtained were aligned with reference sequences of the seven genotypes available in GenBank. Phylogenetic analysis was performed in MEGA 7 software using the maximum-likelihood method with nucleotide substitution model GTR+G+I selected as the best fit for the input multiple sequence alignment. Bootstrap values using 1000 replicates were accessed to test and estimate the confidence of evolutionary relatedness between HPgV-1 sequences.
2.4 Statistical analysis
HPgV positivity and independent risk factors were transformed into categorical variables and Fisher's exact test was used to examine possible relationships between them. Mann-Whitney test was used for nonparametric analyses. P< .05 were considered significant. Statistical analyses were performed using RStudio Desktop (version 1.2.1335).
3 RESULTS
We analyzed the HPgV-1 RNA presence among 257 serum samples collected from 61 kidney transplant recipients (24 females and 37 males), during the first year posttransplant. All organs but one were from deceased donors and the most common primary diseases observed were chronic glomerulonephritis (23%), systemic arterial hypertension (21.3%), and systemic arterial hypertension associated with diabetes mellitus 2 (13.1%). Table 1 shows the characteristics of the 61 patients according to their HPgV-1-RNA status. HPgV-1 was found in 36.1% (22/61) of patients, at least in one blood collection and there was no coinfection with HBV, HCV, or HIV. Statistical analysis revealed a correlation between HPgV-1 and female sex (P = .01) but not with other demographic and clinic variables.
| All patients (%) | HPgV-1 negative (%) | HPgV-1 positive (%) | P valuea | |
|---|---|---|---|---|
| Patients, n | 61 (100) | 39 (63.9) | 22 (36.1) | |
| Age | ||||
| 20-35 | 14 (23) | 9 (64.3) | 5 (35.7) | |
| 36-50 | 31 (50.8) | 21 (67.7) | 10 (32.3) | .739 |
| ≥51 | 16 (26.2) | 9 (56.2) | 7 (43.8) | |
| Sex | ||||
| Male (%) | 37 (60.7) | 28 (75.7) | 9 (24.3) | .01 |
| Female (%) | 24 (39.3) | 11 (45.8) | 13 (54.2) | |
| Primary disease (%) | ||||
| Chronic glomerulonephritis | 14 (23) | 11 (78.6) | 3 (21.4) | |
| Systemic arterial hypertension | 13 (21.3) | 8 (61.5) | 5 (38.5) | .23 |
| Systemic arterial hypertension + diabetes mellitus 2 | 8 (13.1) | 6 (75) | 2 (25) | |
| Otherb | 24 (39.3) | 14 (58.3) | 10 (41.7) | |
| Unknown | 2 (3.3) | … | 2 (100) | |
| HLA match, n (%) | ||||
| 0 | 1 (1.6) | 1 (100) | 0 | .17 |
| 1-3 | 51 (83.6) | 30 (58.8) | 21 (41.2) | |
| 4-6 | 9 (14.8) | 8 (88.8) | 1 (11.2) | |
| Induction therapy | ||||
| Thymoglobulin, n (%) | 41 (67.2) | 29 (70.7) | 12 (29.3) | .15 |
| Basiliximab, n (%) | 20 (32.8) | 10 (50) | 10 (50) | |
| Immunosuppressive drugs, n (%) | ||||
| Tacrolimus + prednisone + mycophenolate mofetil | 31 (50.8) | 17 (54.8) | 14 (45.2) | .18 |
| Tacrolimus + prednisone + mTOR inhibitors | 28 (45.9) | 21 (75) | 7 (25) | .11 |
| Others | 2 (3.3) | 1 (50) | 1 (50) | .99 |
| Delayed graft function | .54 | |||
| No | 45 (73.8) | 30 (66.7) | 15 (44.3) | |
| Yes | 16 (26.2) | 9 (56.2) | 7 (43.8) | |
| Graft dysfunction, n (%) | .09 | |||
| No | 36 (60.0) | 20 (55.6) | 16 (44.4) | |
| Yes | 24 (40.0) | 19 (79.2) | 5 (20.8) | |
| Rejection | .77 | |||
| No | 43 (70.5) | 28 (65.1) | 15 (34.9) | |
| Yes | 18 (29.5) | 11 (61.1) | 7 (38.9) | |
| Graft loss | .73 | |||
| No | 51 (83.6) | 32 (62.7) | 19 (37.3) | |
| Yes | 10 (16.4) | 7 (70) | 3 (30) | |
| Receptor median age, y | 44 | 44 | 43.5 | .927 |
| Donor median age, y | 51 | … | … |
- Note: Bold value indicate statistically significant variable analyzed.
- Abbreviations: HLA, human leukocyte antigen; HPgV-1, human pegivirus-1; mTOR, mammalian targetof rapamycin.
- a P value from Fisher's exact test or Mann-Whitney test.
- b Other primary diseases that individually account for less than 5% of cases.
Detectable viral RNA was observed in all blood collections in 50% (11/22) of HPgV-1 infected individuals. Of these, one patient died after two samplings. In five renal recipients, samples comprising the whole period of follow up (12 months) were constantly viremic. For the remaining five patients which RNA HPgV-1 was detected in all samples, but blood collections were performed only for 6 months after transplantation.
Viremia was detected in, at least, one collection point for the remaining 11 subjects who tested positive. For all but one of these patients, HPgV-1 RNA was not detected in the first month posttransplantation. Viral RNA was detected in 10 patients after 3 months. Of these, one subject (ID 41) had two consecutive positive samples (months 3 and 6) and one participant (ID 9) had HPgV-1 RNA detected only in the last sample, collected after 12 months posttransplantation, suggesting a newly acquired HPgV-1 infection. Interestingly, a fluctuation in viral RNA presence was noted in one patient (ID 5), who tested positive in samples collected in months 6 and 12, with a negative sample between this interval (Table 2).
| Months posttransplantation HPgV-1 RNA/genotype | |||||
|---|---|---|---|---|---|
| Patient ID | 1 | 3 | 6 | 9 | 12 |
| 5 | − | − | +/2b | − | +/2a |
| 9 | − | − | − | − | +/1 |
| 12 | − | − | +/2b | − | − |
| 18 | − | − | +/3 | − | − |
| 40 | − | +/2b | − | NA | NA |
| 41 | − | +/2b | +/und | − | NA |
| 54 | − | +/2b | − | − | − |
| 55 | − | +/und | − | − | NA |
| 63 | − | +/2b | − | − | − |
| 78 | − | − | +/2b | − | − |
| 99 | +/2b | − | − | − | − |
- Abbreviations: HPgV-1, human pegivirus-1; ID, identification; NA, not available; und, undetermined.
Genotypes were determined by phylogenetic analysis of the 5′UTR of HPgV-1 samples obtained from 21 of 22 positive individuals (Figure 1). Due to the low quality of the nucleotide sequencing, a sample from one individual was not suitable for performing phylogenetic analysis. Seventeen patients were infected by genotype 2, corresponding to 80.9% (17/21) of the total. Subtypes 2a and 2b were identified in 17.6% (3/17) and 76.5% (13/17) of samples, respectively. Interestingly, for one patient (ID 5) (1/17; 5.9%), both subtypes were found in different time points of blood collections. Genotype 3 was identified in two individuals (9.5%) and, finally, genotypes 1 and 5 were also identified into one (4.8%) sample each. HPgV-1 sequences obtained here were deposited in GenBank under accession numbers: MK624940 to MK624947 and MK624949 to MK624963.
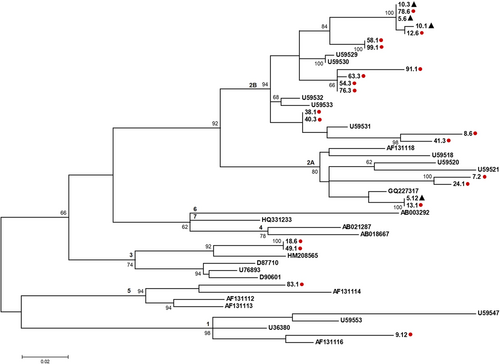
Phylogenetic analysis indicated that sequences that were classified as subtype 2a formed two separated groups (Figure 1). Subtype 2a reference sequences were from Brazil, South Africa, Europe, and USA. Sequences 5.12 and 13.1 were identical and clustered together with a previously described Brazilian sequence. The other 2a sequences, 7.2 and 24.1, were closely related to a sequence reported in Europe. The sequences obtained from kidney recipients, classified as subtype 2b, clustered together in small groups, randomly spread in the whole 2b cluster, along with sequences from Europe and USA. Two sequences were identified as genotype 3 and they clustered together with a Brazilian one, separated from Asiatic sequences from Japan and Taiwan. Here, one sequence was subscribed as HPgV-1 genotype 1 and one sequence as HPgV-1 genotype 5. This latter sequence clustered together with reference sequence from South Africa, whereas HPgV-1 genotype 1 sequence did not cluster together with reference sequences, forming a separated lineage inside genotype 1 (Figure 1).
Nucleotide sequences analyses of 19 patients with multiple collection points indicated that HPgV-1 sequence remained identical during all the evaluated periods. An identical nucleotide sequence was also observed between different recipients. Patients ID 5, ID 10, and ID 78, shared the same sequence, as well as (a) patient ID 58 and ID 99; (b) patient ID 54 and ID 76; (c) patient ID 38 and ID 40; (d) patient ID 5 and ID 13; and (e) patient ID 18 and ID 49 (Figure 1). For patient ID 10, the substitution of two nucleotides was observed between the first collection point (1 month after the transplant; ID 10.1) and the second one, 2 months later (ID 10.3). These substitutions were maintained in the subsequent three points. Patient ID 5, positive for the presence of HPgV-1 RNA in only two collection points (6 months; ID 5.6, and 12 months; ID 5.12, after transplant, with a negative result in the further points), presented HPgV-1 sequences classified in different subtypes. Sample collected 6 months after the transplant was characterized as 2b while a 12-month sample indicated infection with subtype 2a.
4 DISCUSSION
Kidney transplantation is the best treatment option for patients with end-stage renal disease, improving quality of life and contributing to a better prognosis for these patients. Despite the development of more effective immunosuppressive agents had led to a significant reduction of acute rejection and improved graft survival, its administration is also associated with various posttransplant complications, such as infectious diseases.3, 16
In the present study, we determined the prevalence and genotypic characteristics of HPgV-1 in kidney transplant recipients. Until now, although HPgV-1 has been considered a nonpathogenic agent, its activity reducing immune system activation has been investigated17, 18 and how this mechanism could impact organ transplantation outcome is still poorly understood.19-21
The prevalence of HPgV-1 viremia among renal transplant patients was 36.1%, much higher than the frequency found in a previous Brazilian study, where HPgV-1 RNA was detected in 16.7% among kidney recipients.21 This discrepancy might be explained by the low number of transplant patients evaluated in the former study (n = 12), different locality of the study population, and/or methodology used to perform the molecular tests. This rate was also much higher than found in studies carried on developed countries as the USA, France, and Italy, which ranged from 17% to 26%.19, 22-24 Regarding developing countries, HPgV-1 prevalence in renal recipients was still higher than the demonstrated here, affecting 43% individuals in Thailand,25 41.2% in South Africa,26 42% in Turkey,20 and 52.9% in India.27 Although HPgV-1-RNA detection by RT-PCR had targeted different genomic region (5′UTR, NS3, and NS5) in these studies, which could interfere in results due to inner sensitivity and specificity of each employed methodology, the difference in detection rates emphasizes the high prevalence of viral RNA in low-income countries, mirroring what is observed for other infectious diseases with similar modes of transmission, such as HIV and HCV.
There was no statistical association between HPgV-1 viremia and age, primary disease, human leukocyte antigen match, induction therapy, immunosuppressive drugs, rejection, and graft loss events. However, for graft dysfunction, a P value very close to the cut off for statistical significance was observed when testing the association with HPgV-1 infection. Further analysis with a larger sampling size might shed a light if the presence of HPgV-1 would be a negative factor for graft dysfunction. As expected, we confirmed the high frequency of chronic glomerulonephritis systemic arterial hypertension and diabetes mellitus 2 considered the most common comorbidities associated with the development of kidney dysfunction.28, 29 Statistical comparisons between HPgV-1 RNA positive and negative groups indicated that HPgV-1 infection in women was more frequent than in men (P = .01), a result previously reported in a cohort of patients with end-stage renal disease in the USA,24 an association that might be further investigated to suggest a plausible explanation.
In this cohort, 50% (11/22) of seropositive participants were persistently viremic up to 1 year after transplantation, an unexpected result, since it is estimated that HPgV-1 infection persists in approximately 25% of infected individuals and 75% to 80% are able to clear viremia up to 2 years of infection.18 On the basis of this information, at least for our 1-year follow-up research, immunosuppressive status posttransplantation might have interfered in viral clearance or persistence among renal transplant recipients. We observed newly acquired infections in 50% of HPgV-1 positive individuals and 45.5% (10/22) of them cleared the virus a few months after infection. This fast clearance was also observed in a cohort of men who have sex with men followed up for 1 year30 and in recipients of HPgV-1-positive blood, also followed for 1 year after transfusion.31 One patient (4.5%) infection was detected in the last blood collection, 12 months after transplantation, preventing us to observe whether HPgV-1 infection would persist or suppressed.
In Brazil, few studies have evaluated the circulating genotypes of HPgV-1 and the main studied population was blood donors and HIV-infected individuals. Genotypes 1, 2, and 3 were commonly found, but strains from genotype 2 and its subtypes (2a and 2b) constituted the majority and most widely spread in Brazil, ranging from 72% to 98% of sequenced samples.14, 21, 32-38 Our results demonstrate the high prevalence of genotype 2 among kidney recipients in Rio de Janeiro, identified in 80.9% of cases; followed by 9.5% of genotype 3% and 4.8% of genotypes 1 and 5 each. As in previous studies performed in Brazil, subtype 2b was the most prevalent, identified into 76.5% (13/17) of genotype 2 HPgV-1-infected patients. Interestingly, in one patient (ID 5), both subtypes 2a and 2b were found in different points of follow up, with a 6-month interval between blood collections and a negative result during this interval. This may reflect a case of clearance of infection and reinfection, but, unfortunately, the lack of demographic data to support multiple episodes of exposition to HPgV-1 in this individual prevented us to further discuss this fact.
Interestingly, we found one patient infected by HPgV-1 genotype 5. Genotype 5 circulates within central and southern Africa and here we report the first detection of this genotype in Brazil. Further investigation and analysis of a larger genomic fragment might elucidate possible routes of dissemination and entrance of HPgV-1 genotype 5 in Brazil.
Substitutions in nucleotide sequences between different collection points were only observed in one patient (ID 10). In this case, two transition between purines (in positions A318G and G453A, according to the reference sequence U44402) occurred between the first and second collection. These substitutions were maintained in the subsequent three points. This result is in accordance with a previous study analyzing genetic diversity of 5′-UTR region that indicated transitions of one or two nucleotides as the most common substitutions observed in the noncoding genomic sequence.39 The high similarity observed among HPgV-1 sequences infecting different patients and its stability over collection points could be explained by the characteristics of the genomic region analyzed. The choice of the analysis fragment (5′-UTR) was based on the availability of international sequences to enable a proper comparison with the Brazilian sequences. Nonetheless, the short length of the amplified fragment was not informative enough to support a likely chain of transmission between the organ recipients who attended this transplantation center.
In this study, we investigated the frequency of HPgV-1 infection in renal transplant recipients and, despite the high prevalence of viremia found, no significant impact in patient outcome was observed. We observed a high rate of persistence among positive individuals. Larger epidemiologic studies are needed to elucidate whether immunosuppression induced after organ transplantations may increase persistence rates. Viral infections may have an impact on allograft function and may contribute to the risk of rejection, especially considering the current era of potent immunosuppression protocols before and after kidney transplantation. To reduce the risk of such infections, even those viruses considered as nonpathogenic in healthy hosts should be evaluated when monitoring patient outcomes after organ transplantation.
ACKNOWLEDGMENTS
The authors would like to thank the DNA Sequencing Platform FIOCRUZ for performing nucleotide sequencing. This study was supported by the Oswaldo Cruz Institute—Oswaldo Cruz Foundation/FIOCRUZ—Brazil.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Study design: RBV and CCS; data curation: RBV; data analysis: MAPH, FCAM, and CCS; data collection: TCSW, TAM, and DBMC; methodology: FS-R and JGP; project administration: CCS; and writing—original draft: FCAM and CCS. All authors read and approved the manuscript.