The excretion rate and stability of HAAg in human fecal samples after live attenuated hepatitis A vaccination
Abstract
The live attenuated hepatitis A virus vaccine (HA-L) is in routine use in the Chinese national immunization program (NIP). The major disadvantages of HA-L include that theoretically, it may be possible for mutation shifts and secondary infections of the live vaccine viral strain. The aim of this study was to explore variation in the viral strain after vaccination with the HA-L. A total of 1297 fecal samples (including 470 for the 18 to 36-month-old age group, 527 for the 3 to 16-year-old group, and 300 for the 16 years and older group) were collected in the study, and the rate of hepatitis A virus (HAV) positivity in fecal samples was 11.36% (31/273), 11.44% (31/271), 9.70% (26/268), 8.47% (21/248), and 9.70% (23/237) on days 0, 7, 14, 21 and 28, respectively. A total of 77 HAV positive samples were randomly selected for VP1/2A (360 bp, 2218-2577) gene analysis. Phylogenetic trees were then constructed by the neighbor-joining method. Phylogenetic analyses showed that all the isolated HAV strains belonged to sub-genotype IB, which was the same as the vaccine strain. Compared with the vaccine strain, HM-175/7MK-5 (M16632.1), there were only two base mutations discovered, at 2291 and 2568. However, the amino acid mutation analysis showed that those base mutations were synonymous mutations. The isolated HAV strains were genetically stable. This study provides a reference for the safety concern regarding the routine and wide-range use in people older than 18 months.
Highlights
-
The rate of HAV positivity in fecal samples was 11.36%, 11.44%, 9.70%, 8.47% and 9.70% on days 0, 7, 14, 21 and 28, respectively. The HAV strains isolated from HAV positive fecal samples, belonged to sub-genotype IB (vaccine strain). We analyzed VP1/2A (360 bp) regeion and found that there were only two synonymous mutations in isolated HAV strains compared with vaccine strain.
1 INTRODUCTION
Hepatitis A virus (HAV), a member of the genus Hepatovirus in the family Picornaviridae, is one of the most common infectious etiologies of acute hepatitis worldwide.1 The HAV genome is a linear single-stranded RNA, composed of 7500 nucleotides. HAV is composed of 4 capsid proteins (VP1, VP2, VP3, VP4) and seven nonstructural proteins (2A, 2B, 2C, 3A, 3B, 3C, 3D).2-4 The viral proten (VP) 1/2A junction (168 nt) is used to is classified into six genotypes (I-VI), of which genotypes I, II, and III are found in humans.5-7 These are further divided into subgenotypes IA and IB, IIA and IIB, and IIIA and IIIB, respectively.8, 9 Genotype I is the most prevalent worldwide, and subgenotype IA is more common than IB.10
According to the WHO estimated in 2016, 7134 persons died from hepatitis A worldwide.11 HAV is primarily transmitted fecal-orally via contaminated food or water or through close contact with an infected person. With improved sanitation and provisions of the HAV vaccination, the HAV endemicity was declining. HA vaccines have been commercially available since 1990 in China. China is one of few countries worldwide in which both inactivated HA vaccine (HA-I) and live attenuated hepatitis A vaccine (HA-L) are approved for use.12, 13 HA-L is given as a single dose to children older than 18 months.14 Due to the low cost, a single injection dose conferred high protection and increased the duration of immunity through natural boosting of subclinical infection, 27 of the 31 provinces of China enroll the HA-L vaccine into their routine childhood immunization program.15 A former study reported that there were 22 cases of reversion to virulence documented in over 300 million doses of the live attenuated yellow fever (YF- strain 17D) vaccine administered.16 The major disadvantages of HA-L are the potential horizontal transmission due to vaccine viral shedding and the theoretically possible reverse mutation of the live vaccine strains.
The aim of this study was to explore the variation of the viral strain after vaccination with the live attenuated hepatitis A vaccine. Fecal samples from vaccinated participants were screened for the extraction of HAV. Then, VP1/2A regions of HAV were amplified, sequenced and compared to others described and submitted to GenBank, including the sequence of the original vaccine strain. This study provides a reference for the safety concern of the routine and wide-range use of the Chinese national immunization program (NIP).
2 METHODS
2.1 HA-L vaccination and sample collection
Participants involved in this study were divided into three age groups, 18 to 36 months age group, 3 to 16 years old group, 16 years and older group and were administered the HA-L vaccine. The parents or legal guardians of all enrolled participants provided written informed consent before any study procedures. This trial is registered with ClinicalTrials.gov, number NCT02601040.
Fecal samples were collected on 0, 7, 14, 21 and 28 days after HA-L vaccination. Then, 4 g of fecal samples were dissolved in 15 mL 0.01 M phosphate-buffered saline. Fecal supernatant was collected after 4°C centrifugation for 30 minutes at 3500 rpm.
2.2 Detection of HAV antigen
HAV antigen was detected by enzyme-linked immunosorbent assay (ELISA; Kehua Bio-Engineering Company, Shanghai, China) in the supernatants of fecal samples, according to the instructions of the manufacturer.
2.3 Virus isolation
The HAV positive samples were selected to isolate HAV. In total, 100 μL of the fecal supernatant was inoculated into a 25 cm2 bottle with a monolayer of human embryo lung diploid (KMB17) cells. After 2 hours of adsorption, the supernatant was discarded, and the cells were cultured in RPMI 1640 culture medium with 2% serum for 7 days, followed by subculturing two times.
2.4 RNA extraction
Viral RNA was extracted from 140 μL of culture supernatant using the QIAamp viral RNA mini kit (Qiagen, Germany), in accordance with the manufacturer's instructions.
2.5 Primer design
HAV HM-175 strain (GenBank accession number: M14707) was selected to design reverse transcription-polymerase chain reaction (RT-PCR) primers targeting the VP1/2A junction region by primer 5, Forward: ACTGGATGGTTTGGGTGACAA; Reverse: GTCTCCCAGTAAGAACCCCAG.
2.6 RT-PCR
RT-PCR was carried out using TaKaRa PrimeScript One Step RT-PCR Kit Ver2 (TaKaRa Biotechnology, Dalian) with the following protocol: initial reverse transcription at 50°C for 30 minutes; denaturation at 94°C for 2 minutes; 35 cycles of denaturation at 94°C for 30 seconds, annealing at 55°C for 30 seconds, and elongation at 72°C for 1 minute; and a final elongation step at 72°C for 5 minutes. PCR products were confirmed by agarose gel electrophoresis and sent to Sangon Biotech (Shanghai, China) for sequencing.
2.7 Phylogenetic analysis
The sequences from this study were submitted to GenBank for the accession numbers. The analysis of bases and amino acids of the VP1/2A region (360 bp, 2218-2577) of isolated HAV strains was performed by BioEdit. The phylogenetic analysis, based on the VP1/2A genes, was conducted using Molecular Evolutionary Genetics Analysis (MEGA) software version 6.0 (the neighbor-joining method). A total of 77 sequences of isolated strains were deposited into the NCBI GenBank database (http://www.ncbi.nim.nih.gov/GenBank/index.html); samples were randomly selected, including MF774239 to MF774323 and MF776991 to MF777022. The reference viral sequences used to construct the distinct phylogenetic branches were collected from the GenBank sequence database under the accession numbers shown in Table 1.
| Genotypes | Subgenotypes | Strains | Accession numbers |
|---|---|---|---|
| I | Type IA | hd9 | KP177965.1 |
| IT-SCH-00 | AJ505572.1 | ||
| HAJ88-5 | AB258550.1 | ||
| GBM/WT | X75215.1 | ||
| … | K02990.1 | ||
| HAV6 | EU526088.1 | ||
| MB-7/4647 | EU251188.1 | ||
| HA16-0511 | LC191189.1 | ||
| HAV5 | EU131373.1 | ||
| HAV-Arg/06 | HM769724.1 | ||
| FH3 | AB020569.1 | ||
| Type IB | HAV/Hu/2014/TR04 | KT222952.1 | |
| HM-175/7MK-5 | M16632.1 | ||
| HAF-203 | AF268396.1 | ||
| HAV_NL_2010000906 | KM261588.1 | ||
| NCBI Reference Sequence: | NC_001489.1 | ||
| LA-1 | AF314208.1 | ||
| … | M20273.1 | ||
| HM-175 (wild type) | M14707.1 | ||
| II | Type IIA | CF53/Berne | AY644676.1 |
| Type IIB | SLF88 | AY644670.1 | |
| III | Type IIIA | NOR-21 | AJ299464.3 |
| HAV/GIU/BA/2012/IT | KF706410.1 | ||
| Type IIIB | HA-JNG06-90F | AB258387.1 | |
| HAJ85-1 | AB279735.1 |
3 RESULTS
3.1 Diagnosis of HAV
A total of 1365 fecal samples were collected on days 0, 7, 14, 21 and 28 after vaccination. Among them, there were 31 HAs-Ag positive participants at the first collection (day 0), 31 HAs-Ag positive participants at the second collection (day 7), 26 at the third collection (day14), 21 at the fourth collection (day 21) and 23 at the last collection (day 28). The average rate of positive HAAg in this experiment was 11.36% (31/273) at the first collection, 11.44% (31/271) at the second collection, 9.70% (26/268) at the third collection, 8.47% (21/248) at the fourth collection and 9.70% (23/237) at the last collection. Since the number of positive participants was descending, the comparison among different groups was insignificant. The results are shown in Table 2.
| Participants (Group) | The rate of positive HAs-Ag in fecal samples | ||||
|---|---|---|---|---|---|
| The first time | The second time | The third time | The fourth time | The fifth time | |
| 18-36-mo age group | 4.60% (4/87) | 9.52% (10/105) | 6.00% (6/100) | 5.38% (5/93) | 5.88% (5/85) |
| 3-16-y-old group | 15.13% (18/119) | 14.42% (15/104) | 11.32% (12/106) | 14.14% (14/99) | 14.14% (14/99) |
| 16 y and older group (including 16-y-olds) | 13.43% (9/67) | 9.68% (6/62) | 12.90% (8/62) | 3.57% (2/56) | 7.55% (4/53) |
| Total | 11.36% (31/273) | 11.44% (31/271) | 9.70% (26/268) | 8.47% (21/248) | 9.70% (23/237) |
3.2 Phylogenetic analysis and population dynamics of HAV strains
To amplify the virus, 132 positive samples were inoculated into KMB17 cells. After 26 days of culture, samples were randomly selected for HAV RNA sequencing. A total of 77 samples were randomly selected, including 21 in the 18 to 36 month group, 24 in the 3 to 16-year-old group, and 32 in the 16-year-old and above group. The VP1/2A polyprotein genes of HAV isolated strains in this study were determined and analyzed by aligning their sequences with 25 other representatives HAV strains of diverse origins retrieved from GenBank (Figure 1). According to the neighbor-joining tree of the 360-nt VP1/2A sequence, all the sequences in this study belonged to genotype IB. The HA-L vaccine produced by the H2 strain was administered in this study, so the sequence of the attenuated strain, HM-175 (M16632.1), was used as a standard reference strain. Phylogenetic analysis showed that VP1/2A from all isolated strains belonged to or was close to the cluster of HM-175. The cluster that contained HM-175 was defined as cluster 1 and accounted for 62.00% (48/77) of all isolates.
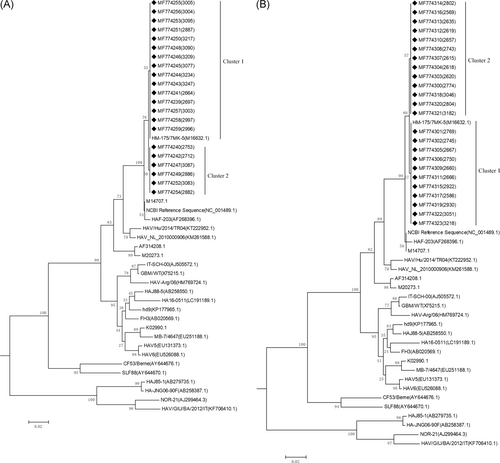
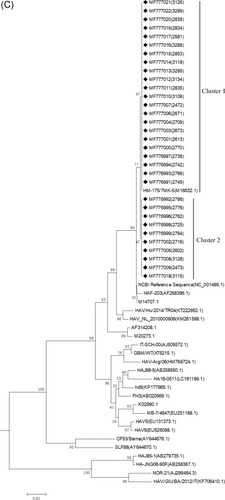
3.3 Analysis of base and amino acid mutations of VP1/2A region in isolated HAV strains
Compared with the vaccine strain, HM-175/7MK-5 (M16632.1), there were only one base mutations (T→A) discovered at position 351. The position of this single-base mutation was at 2568 in the whole coding sequence, which belongs to cluster 2 (Figure 2). The amino acid mutation analysis showed the base mutation was synonymous mutation(see the red box in Figure 2). The HAV isolated strains in this study were genetically stable in the VP1/2A region.

4 DISCUSSION
Genotype I is the most prevalent worldwide and is often found in North and South America, Europe and Asian countries, including China, Indonesia, and Japan.17, 18 HA-L originated from H2 or LA-1 strains was approved for use in China; both viral strains belong to Genotype I. In this study, HA-L originating from the H2 strain was selected for administration. In our study, the excretion of HAAg was investigated in stool samples on days 0, 7, 14, 21, and 28 after vaccination of HA-L in the three age groups (18-36 months, 3-16 years old and 16 years and older). All the 77 isolated HAV strians from the fecal samples of vaccinators were classified into genotypes IB.
In China, only three developed cities and one flourishing province use HA-I for the routine childhood immunization program. Nevertheless, due to the low cost and simple immunization procedure (ie, a single dose), the other 92% of individuals (16 million annual births) have been immunized with HA-L since 2008 in other provinces in China.19, 20 The major disadvantages of HA-L include the theoretical possibility of those mutation shifts of the live vaccine strain and secondary infections. Our results showed that approximately 25% of the participants tested were positive for HAAg in fecal samples within 28 days after administration. Despite individual differences in vaccination, the excretion of HAAg among different age groups was insignificant. In other words, the vaccine virus was copied and excreted actively in the immunized body. Therefore, based on the large-scale use of HA-L, secondary infection and the possibility of mutational shifts of the live vaccine virus cannot be neglected. More studies are needed to explore the possibility of secondary infections and mutational shifts of the live vaccine virus in the field. One of the safety concerns for HA-L is regarding the mutation shifts of the live vaccine strain. In this study, there were no nonsynonymous mutations detected in the part of the VP1/2A region (360 nt) of all HAV isolated strains. More experiments are needed to investigate the possible mutations of other coding regions of the HA-L strain.
With the decreasing circulation of HAV in the population as a result of mass HA vaccination and improvement of sanitation, the chance of subclinical HAV infection is becoming rarer, and one cannot rely on the booster effect of subclinical infections for a longer term of vaccine protection. A former study reported that HA-L is more genetically stable, more immunogenic in animal studies and non-transmissible by the oral route.21, 22 This study provides a reference for the safety concern of the routine and wide-range use in people that are older than 18 months.
ACKNOWLEDGMENTS
We thank the Institute of Medical Biology, Chinese Academy of Medical Sciences, and Peking Union Medical College for providing inactivated vaccines (Weisairuian) and live attenuated vaccines (Weisairuiji). Many thanks to all the investigators kindly working on the trial from Jiangsu Provincial Center of Disease Control and Prevention. We thank Drs. Penghua Wang in the Department of Microbiology & Immunology, School of Medicine, New York Medical College, and Nigel W. Fraser, Prof. Emeritus, Dept of Microbiology, Perelman School of Medicine, University of Pennsylvania for proofreading and reviewing our manuscript. This study was supported by the National “Twelfth Five Year” “major new drug discovery” technology major projects (Grant No. 2012ZX09104-302), the Jointly Supported Foundation of the National Project in Yunnan Province (2013GA018), the CAMS Initiative for Innovative Medicine (CAMS-I2M) (Grant No. 2017-I2M-2-006), the National Natural Science Foundation of China (Grant No. 81171946), and the Natural Science Foundation of Yunnan Province (Grant No. 2016FA029).
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
XW wrote the article and contributed to data acquisition and phylogenetic analysis. XW and YP contributed to virus isolation. YP and JC contributed to RNA extraction and RT-PCR. JL, JX, and CY contributed to the detection of HAV antigen and sample collection. YZ, LQ, SW, SH, and YL contributed to data acquisition. QS designed the study and reviewed the manuscript for its intellectual content.
ETHICS STATEMENT
The study protocol was approved by the Institutional Ethics Committee (Institute of Medical Biology, Chinese Academy of Medical Sciences, and Peking Union Medical College) and was in accordance with the Declaration of Helsinki for Human Research of 1974 (last modified in 2000).