HIV-1 genetic diversity and transmitted drug resistance among newly diagnosed HIV-1 individuals in Jiangmen, China
Abstract
Jiangmen is one of the Guangdong-Hong Kong-Macao Greater Bay Areas with frequent commercial intercourse, which is responsible for human immunodeficiency virus type 1 (HIV-1) rapid circulation and genetic evolution for recent years. As a novel HIV-1 second-generation recombinant was previously reported in Jiangmen but the systematic molecular epidemiological investigation was still unknown. A retrospective study on HIV-1 genotypic characteristics and the emergence of transmitted drug resistance in this region was necessary. A total of 224 newly diagnosed HIV-positive cases were randomly selected in Jiangmen City of Guangdong Province between 2018 and 2019. The partial gag (1080 bp), pol (840 bp), and env (460 bp) genes were amplified using nested polymerase chain reaction followed by sequencing. The phylogenetic and recombination analysis as well as HIV-1 drug resistance were performed to surveillance. Sexual transmission was determined to be the major risk factor in Jiangmen. Phylogenetic analysis detected the genotypic distribution as follows: CRF01_AE (36.65%,70 of 191), CRF07_BC (32.46%, 62 of 191), CRF08_BC (4.71%, 9 of 191), CRF55_01B (5.24%, 10 of 191), CRF59_01B (3.14%, 6 of 191), subtype B (4.71%, 9 of 191), subtype C (1.05%, 2 of 191) as well as unique recombinant forms (12.04%, 23 of 191) consisted of seven recombinant patterns, which originated from multiple regions of China. Low-level prevalence of Surveillance Drug Resistance Mutations (2.1%) were predicted but drug-resistant mutations showed at a high level (15.4%) especially mutations in RT gene at position 179 were found to be the most frequent in the therapy-naïve population. Our study highlighted the critical importance of monitoring the emerge of recombinant strains among newly diagnosed HIV-1 individuals along with drug resistance regularly to prevent multi-channel introduction and breakout of new HIV strains.
Highlights
-
Our study highlighted the critical importance of monitoring the emerge of recombinant strains among newly diagnosed HIV-1 individuals along with drug resistance regularly to prevent multi-channel introduction and breakout of new HIV strains.
1 INTRODUCTION
It was estimated that around 770 000 people died of the human immunodeficiency virus (HIV)-related illnesses globally in 2018.1 The HIV/AIDS epidemic has still posed a severe public health concern around the world in terms of its treatment, management, and prevention. It was pointed that 849 602 people had infected with HIV in China by the end of September 2018.2 Since National Free Antiretroviral Treatment Program has been made available free of charge to HIV-1-infected patients widely since 2003,3 it should be noted that the resistant strains carrying drug-resistant mutations were observed in treatment-naïve patients gradually, getting more chances to generate drug-resistant strains potentially or directly.4 Transmitted drug resistance (TDR) has become a major obstacle wrecking the effect of antiretroviral therapy (ART) among newly diagnosed individuals. Further surveillance and intervention might be necessary to reduce the risk of transmission on drug-resistant viruses.
Owing to the high genetic variation of HIV-1, extensive genetic diversity and ceaseless occurrence of new recombinants were developing, which could easily accelerate the generation and evolution of HIV-1 variants.5 It had reported that there were at least 21 circulating recombinant forms (CRFs) and 8 subtypes in China since 2013.6 Particularly, some novel CRFs and unique recombinant forms (URFs) comprised of different genotype gene segments were identified in succession for recent years, especially some specific areas with high level of HIV-1 infection and genetic diversity,4, 7 leading much more challenges in the prevention and control of HIV epidemics in China.
Located in Chinese southern mainland, Guangdong is one of the most opening areas with a wide range of floating population for business or tourism, which was lying the fifth place of the highest proportion of HIV/AIDS cases nationwide.8 Notably, some novel HIV-1 recombinants like CRF55_01B/CRF59_01B had been recently reported in some prefectures such as Shenzhen and Dongguan,9, 10 suggesting a complicated epidemic trend in Guangdong. Numerous research had investigated molecular epidemiology of HIV-1 focused on first-tier cities in China but it was not enough to reflect the comprehensive genetic diversity of HIV-1 infection. Surveillance for emerging HIV variants on a periodic basis in non-first-tier cities or rural areas was equally warranted owing to the dynamic change of HIV-1 subtypes and recombinants.
As a famous hometown of overseas neighboring Hong Kong and Macao, Jiangmen is one of the Guangdong-Hong Kong-Macao Greater Bay Areas with frequent commercial intercourse, which is responsible for HIV-1 rapid circulation and genetic evolution. Moreover, a novel near full-length HIV-1 recombinant strain of CRF07_BC/CRF55_01B (labeled as MK697709) was identified as a second-generation recombinant in 2018 on our previous research,11 we suspected that there might be a coexistence of multiple genotypes in Jiangmen but a systematic genetic characteristic has been insufficient yet. Thus, we aimed to conduct a comprehensive molecular epidemiological investigation on the distribution of HIV-1 genotypes and recombinants as well as assess drug resistance of HIV-1 variants among newly diagnosed HIV-1 positive before receiving ART in Jiangmen lately, which could provide scientific information on the development of effective control of HIV-1 transmission and suitable prevention strategies.
2 MATERIALS AND METHODS
2.1 Participants and specimens
A retrospective study was conducted between February 2018 and May 2019. Newly diagnosed HIV-infected individuals without receiving ART were randomly recruited from the local HIV/AIDS sentinel surveillance sites in Jiangmen, Guangdong Province, China, with informed consents and questionnaires (containing sex, age, occupation and marital status, educational level, mode of transmission, etc). Their plasma samples were collected by professional staff and then used immediately for routine blood count and CD4+T cell count measurements. This study was approved by Medical Ethics Committee of Jiangmen Center for Disease Control and Prevention. The epidemiologic background was collected through specific epidemiological investigation. CD4 cells were calculated using flow cytometry (San Jose, CA).
2.2 Viral RNA extraction, gene amplification, and sequencing
Viral RNAs were extracted from 200 μL of plasma with high pure viral RNA kit (Roche, Basel, Switzerland) according to the manufacturer's instructions and then subjected to nested polymerase chain reaction (PCR) after reverse transcription of HIV-1 gag gene (HXB2:781-1861), pol gene (HXB2:2390-3230) and env gene (HXB2:7840-8300) for HIV-1 genotyping as described in the previous research.12, 13 Positive PCR products were sent to Beijing Liuhe Huada Gene Technology Company (BGI, Beijing, China) for Sanger sequencing after purification.
2.3 Genotyping and phylogenetic analysis of HIV-1 sequences
All sequenced fragments in gag, pol, and env gene regions were amended using BioEdit 7.0 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). The online Quality-Control software (https://www.hiv.lanl.gov/content/sequence/QC/index.html) was performed to check for potential errors. HIV-1 genotypes were identified using HIV BLAST on the Los Alamos HIV Database (https://www.hiv.lanl.gov/content/sequence/BASIC_BLAST/basic_blast.html) and further confirmed by phylogenetic analysis. To demonstrate potential inter-subtype mosaicism, sequences were analyzed using bootscanning and informative site performed by SimPlot 3.5.1 software (S. Ray, Johns Hopkins University, Baltimore, MD).14 The HIV-1 subtypes of each sample were determined by combining the results of the three gene regions. If one of the three gene regions was unavailable, the genotype was designated based on the other two genes. Samples with different genotypic identification from the gag, pol, and env regions were considered as URF, which were labeled by the sequence of gag/pol/env subtype designations. For phylogenetic analysis, all our nucleotide sequences were aligned separately by the ClustalW program implemented in BioEdit 7.0 software. The main reference sequences were obtained from the Los Alamos HIV sequence database (http://www.hiv.lanl.gov), including the major HIV-1 subtypes/CRFs in China. Phylogenetic tree analyses were constructed using the neighbor-joining method based on the Kimura two-parameter model implemented in MEGA X.15 The reliability of topologies was tested by bootstrapping analysis with 1000 replicates. The gene sequences were deposited to GenBank as accession numbers of MN966048- MN966219 for the HIV-1 gag p17–p24 gene region, MN966221- MN966408 for the HIV-1 pol gene region, and MN965851-MN966047 for the HIV-1 env gp41 gene region.
2.4 Drug resistance analysis
The sets of partial pol genes covering most of the protease and the entire RT region were submitted to the online analysis interface of the Stanford HIV Drug Resistance Database to detect Drug Resistance Mutation (DRM) and antiretroviral susceptibility. The surveillance of Drug Resistance Mutations (SDRMs) was assessed based on the World Health Organization 2009 list of mutations.16
2.5 Statistical analyses
All of the social demographic data were doubled entered into Microsoft Excel 2010(Microsoft; Redmond, WA). The categorical variables were conducted as absolute values and percentages. Categorical variables and charts were performed using GraphPad Prism 6 software package.
3 RESULTS
3.1 Demographic characteristics of study subjects
Out of 224 samples from local sentinel surveillance sites in Jiangmen, 557 sequences of HIV-1 were successfully amplified and obtained, including 172 samples in the p17-p24 (gag) regions, 197 samples in the gp41 (env) region and 188 samples in the pol region. Based on sequence analysis of 2 or 3 distinct genes, a total of 191 subjects generated interpretable data set and the demographic and epidemiologic data were summarized in Table 1. Among the 191 samples, the majority of the subjects (70.68%, 135 of 191) were males and people registered their residence locally in Jiangmen were 94.76% (181 of 191) and 1.57% (3 of 191) came from other cities in Guangdong Province, and 3.66% (7 of 191) were from other provinces. The mean age of the participants was 46.02 ± 14.53 years. And 92.67% (177 of 191) of participants were of Han ethnicity and minority ethnicities only accounted for 3.66% (7 of 191), including Zhuang, Korean, Wa, Tujia and Buyi. Of all subjects, 24.84% (46 of 191) were single, 44.50% (85 of 191) were married to women, and 27.75% (53 of 191) were divorced or widowed. In terms of educational level, 4.71% (9 of 191) of them had received junior college-level or higher education degrees while 91.62% (175 of 191) had received high school education or less. As for occupation, unemployed or retired individuals contributed to the largest part (47.64%, 91 of 191) and the farmers ranked the second (19.37%, 37 of 191). The following risk groups were identified: homosexual (17.28%, 33 of 191); heterosexuals (73.30%, 140 of 191); injecting drug users (4.71%, 9 of 191); and unknown (4.71%, 9 of 191). The median CD4+T Cell Count was 213 cells/µL (range, 3-1286 cells/µL).
| Characteristics | No. of genotyped subjects,a N | Percentage of genotyped subjects, % |
|---|---|---|
| Total | 191 | 100.00 |
| Sex | ||
| Male | 135 | 70.68 |
| Female | 51 | 26.70 |
| Unknown | 5 | 2.62 |
| Permanent residence | ||
| Jiangmen City | 181 | 94.76 |
| Other Cities in Guangdong Province | 3 | 1.57 |
| Other Provinces | 7 | 3.66 |
| Age | ||
| <26 | 14 | 7.33 |
| 26-45 | 74 | 38.74 |
| 46-60 | 63 | 32.98 |
| >60 | 34 | 17.80 |
| Unknown | 6 | 3.14 |
| Mean ± SDb | 46.02 ± 14.53 | |
| Ethnicity | ||
| Han | 177 | 92.67 |
| Other | 7 | 3.66 |
| Unknown | 7 | 3.66 |
| Marital status | ||
| Unmarried | 46 | 24.84 |
| Married | 85 | 44.50 |
| Divorced/widowed | 53 | 27.75 |
| Unknown | 7 | 3.66 |
| Educational background | ||
| Illiteracy | 5 | 2.62 |
| Primary/middle school | 134 | 70.16 |
| High/secondary school | 36 | 18.85 |
| Junior college or above | 9 | 4.71 |
| Unknown | 7 | 3.66 |
| Occupation | ||
| Worker | 19 | 9.95 |
| Farmer | 37 | 19.37 |
| Commercial service | 10 | 5.24 |
| Housekeeper | 6 | 3.14 |
| Cadres, teachers and doctors | 4 | 2.09 |
| Unemployed/retired | 91 | 47.64 |
| Others | 11 | 5.76 |
| Unknown | 13 | 6.81 |
| Risk group | ||
| Homosexual | 33 | 17.28 |
| Heterosexual | 140 | 73.30 |
| Injecting drug user | 9 | 4.71 |
| Unknown | 9 | 4.71 |
| CD4+T cell count, cells/µL | ||
| >350 | 46 | 24.08 |
| 200-350 | 46 | 24.08 |
| <200 | 84 | 43.98 |
| Unknown | 15 | 7.85 |
| Median | 213 | |
- a No. of genotyped subjects: the HIV-1 genotype of 191 samples was determined based on at least two fragments of gag, pol, and env sequences and 191 samples were used in the following analyses.
- b SD, standard deviation.
3.2 HIV-1 genotype distribution and identification of URFs
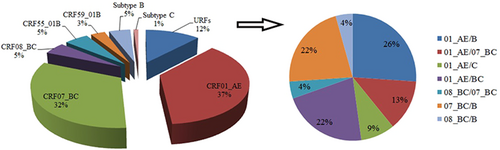
For each sample, 172 gag genes (1080 bp, encoding portions of p17 and p24), 197 env genes (460 bp, encoding the gp41 region), and 188 pol gene (840 bp, encoding portions of protease and reverse transcriptase) were confirmed for genotyping by the neighbor-joining (NJ) trees using a reference set of sequences representing subtypes A-D, F-H, J, K, and common CRFs identified in China from Los Alamos database (http://www.hiv.lanl.gov), shown in Figures 1-3, respectively. By combining the results of phylogenetic tree analyses of three genes, a total of 191 HIV-1 samples with at least two gene regions available were analyzed for HIV-1 genotypes distribution as follows: CRF01_AE (36.65%, 70 of 191), CRF07_BC(32.46%, 62 of 191), CRF08_BC (4.71%, 9 of 191), CRF55_01B (5.24%, 10 of 191), CRF59_01B (3.14%, 6 of 191), Subtype B (4.71%, 9 of 191), Subtype C (1.05%, 2 of 191), and unique recombinant forms (URFs) (12.04%, 23 of 191). Strikingly, the frequency of URFs (12.04%) composed of different genotype segments ranked the third frequent genotype. Confirmed by Simplot software and phylogenetic tree analyses, 23 URFs were distinguished with seven recombinant patterns of gag, pol, and env genes (Figure 4). CRF01_AE/B was the most common recombinant form, accounting for 26.09% (6 of 23), followed by CRF07_BC/B and CRF01_AE/BC, accounting for 21.74% (5 of 23) respectively. And the other four discrete URFs included CRF01_AE/CRF07_BC (13.04%, 3 of 23), CRF01_AE/C (8.70%, 2 of 23), CRF08_BC/CRF07_BC (4.35%, 1 of 23) and CRF08_BC/B (4.35%, 1 of 23). Interestingly, 69.57% (16 of 23) of URFs harbored CRF01_AE gene fragments, suggesting CRF01_AE strain had made a vital contribution with respect to generating new recombinants. Furthermore, there were two inter-subtype recombinant strains (JM.TS016 and JM.jm48) consisting of both CRF01_AE and CRF07_BC gene fragments in the gag region, which had the similar recombinant breakpoint at the overlapping site (Figure S1). The two samples also had a strong connection with URF01BC strain (MH635824) from Shenzhen supported by a high bootstrap value (99%). All of these recombinant subjects were infected through sexual contact (16 heterosexuals, 7 homosexuals).
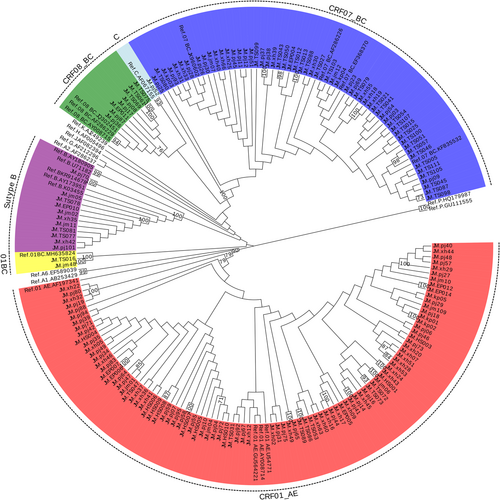
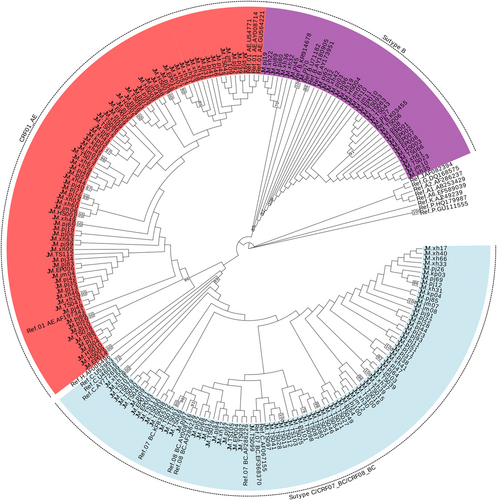
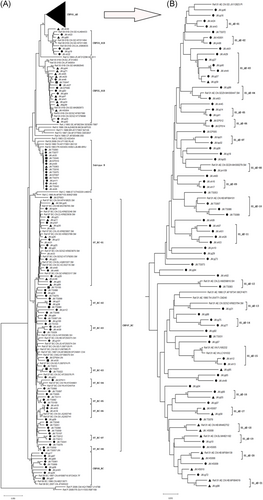
3.3 Genetic relatedness of HIV-1 strains among the prevalent genotype
To investigate the potential origins of the dominant HIV-1 genotype in Jiangmen district with those from other reports, further phylogenetic analysis was carried out. All reference sequences contained were isolated from other cities in Guangdong and different provinces in China as well as some surrounding Southeast Asian countries.188 HIV pol sequences in this study were added to the alignment for the construct of the phylogenetic tree. As shown in the NJ phylogenetic trees of pol genes (Figure 3), CRF01_AE constituted a very large cluster containing 74 study sequences (9 MSM sequences, 60 heterosexual sequences, and 1 infected through IDU) and multiple discrete local clusters within Jiangmen were identified. Some subclusters contained sequences from other cities of Guangdong such as Shenzhen (01_AE-13) and Zhuhai (01_AE-04,08) while few sequences had high geographical linkages with the reference sequences from other provinces including Hebei (01_AE-10,18,20,21), Beijing (01_AE-19), and Zhejiang (01_AE-12). In addition, two specimens were implicated into a cluster with strains from Vietnam (01_AE-15) supported by a high bootstrap value (95%). Moreover, 11 clusters were identified in the phylogenetic trees only contained local heterosexual or homosexual sequences. In the clade of CRF07_BC, a unique cluster (07_BC-01) was formed closely in the MSM population from different provinces across China, clustering with nearly all (6 of 7) MSM isolates in Jiangmen. Besides, 14 heterosexual sequences in the region were also placed in this subcluster. But the remaining CRF07_BC strains intermixed with those through heterosexual and IDU were distinct from the above strains, some of which had strong connections with isolates from Yunnan (07_BC-04) and Guangxi (07_BC-06). Unlike the two prevalent CRFs, two small CRF01B clusters including CRF55_01B and CRF59_01B were firstly detected in Jiangmen and CRF55_01B were potentially related to the other stains from Guangdong. They were both observed between MSM and heterosexuals in Jiangmen.
3.4 Prevalence of TDR-related mutations
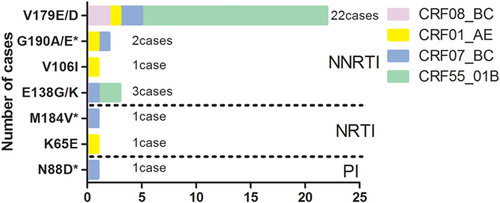
According to Surveillance Drug Resistance Mutations (SDRMs) listed by WHO, 188 pol genes obtained from the therapy-naïve population were sequenced for HIV-1 drug resistance mutations and 2.1% (4 of 188) of them were identified with SDRMs. A total of 15.4% (29 of 188) of 188 subjects with pol genes were identified to be resistant to at least one antiretroviral drug in the therapy-naïve population, with 13.8% (26 of 188) against non-nucleoside reverse transcriptase inhibitors (NNRTIs), 1.1% (2 of 188) against nucleoside reverse transcriptase inhibitors (NRTIs), and 0.5% (1 of 188) against protease inhibitors (PI). As shown in Figure 5, V179E/D was found to be the most frequent mutation with 75.9% (22 of 29) among these Drug Resistance Mutations (DRMs) conferring potential low-level resistance to EFV, ETR, NVP, and RPV, followed by E138K/G (10.3%, 3 of 29), which was associated with intermediate or low-level resistance to RPV and potential low-level resistance to other NNRTIs. Other NNRTI-related mutations G190A/E were found in two cases (6.9%), which would incur intermediate-level to high-level resistance to DOR, EFV, ETR, NVP, and RPV. The rest of the mutations were N88D, K65E, M184V, and V106I, which resulted in different-level resistance to antiretroviral drugs, accounting for 3.4% (1 of 29) of each mutation. In addition, DRMs were almost observed in the sexual transmission isolates, in which 58.6% (17 of 29) of them were heterosexuals while 34.5% (10 of 29) were homosexuals. In an aspect of subtype, various CRFs were found among the 29 samples carrying DRMs, the distribution of which were 100.0% in individuals infected with CRF55_01B (17 of 17), followed by CRF07_BC (6 cases), CRF01_AE (4 cases), and CRF08_BC (2 cases). Notably, drug-resistant sites V179D/E were highly proved to appear in this study, including two samples displaying dual-class drug resistance mutations. There were no DRMs observed in pure subtypes.
4 DISCUSSION
The increasing HIV genetic variants had devastating impacts on many fields of the pandemic, including viral pathogenicity, transmission, diagnosis, clinical supervision, treatment and so on.17 Identification of HIV genotype was the point to explore the distribution of HIV-1 infection, which could offer potential clues to the source of transmission. In this cross-sectional study, we provided the first comprehensive HIV-1 molecular epidemiologic investigation on genotype diversity and transmitted drug resistance survey among HIV-1 newly diagnosed and therapy-naïve population in Jiangmen City of Guangdong Province. All of HIV-1 gag, env, and pol genes were analyzed to give the most systematic information on recombination. Five CRFs, two subtypes and seven kinds of URFs were found circulating in Jiangmen district between 2018 and 2019. Phylogenetic analysis revealed that CRF01_AE and CRF07_BC were the dominant genotypes in Jiangmen. In the meantime, CRF59_01B and CRF55_01B were newly identified mainly transmitted in sexual-contact population, which were remarkably common in Guangdong Province recently.18, 19 Compared with the high prevalence of recombinants focusing on MSM population,20-22 our study found that high emergence of multiple URFs were detected both in heterosexuals and homosexuals, which indicated the multiple spread trend of the co-infection among the heterogeneous HIV-1 variants circulating in Jiangmen. Given that the cases of nonmarital heterosexual contact performing actively in Jiangmen for recent years,23 it is noteworthy that the evaluation of risk exposure and intensify prevention for this population should be underscored.
Tracking the origins of HIV epidemics in HIV-1 newly diagnosed population meant a lot for understanding the transmission direction, directness, or common source, which could accordingly acquire clues for behavior intervention among the target population.24 The genetic relatedness could be inferred from the phylogenetic analysis in our study. Some phylogenetic clusters contained the sequences not only from other cities in Guangdong but also from other distant provinces, including Beijing, Hebei, Zhejiang, and even from regions abroad such as Vietnam in CRF01_AE strains, which suggested multi-channel introductions of CRF01_AE strains from more distant areas into Jiangmen. In addition, multiple discrete clusters were also observed in specimens from Jiangmen, which could be deemed as the local endemic clusters. Previous studies have found that a closely supported phylogenetic subcluster of CRF07_BC strains were identified among MSM within the CRF07 radiation.25-27 In current study, we found that CRF07_BC strains covering nearly all MSM subjects locally and other MSM isolates nationwide assembling as a distinct subcluster, which implied CRF07_BC was overspreading out of its original high-risk population through sexual transmission and the lineages of CRF07_BC strains among MSM population might be new spread trend despite geography.28, 29 Notably, something different found in our study was that partly heterosexuals were also involved in this cluster, which hinted that promiscuous behavior contact emerged between MSM and heterosexuals in Jiangmen. Compared with other high-risk populations, most Chinese MSM preferred to be bisexual or coitus with several sex partners without informing their sexual orientation under social and cultural pressure.30 Because some heterosexuals clustered with the MSM population in our study were single or married, we supposed that some heterosexuals may appear to be bisexual or potential MSM. Accordingly, critical monitor and intervention measures should be combined with HIV-1 testing on these new URFs among newly diagnosed population for the sake of restricting rapid evolution of HIV-1 diversity in Jiangmen.
Because high ART has been the only effective preventive measure yet, the surveillance of drug resistance in China was playing a critical role in public health issues owing to drug-associated mutations devastating impacts on ART. Indeed, the estimated prevalence of TDR was considered as one of the integral parts of the management of decreasing the risk of treatment failure among untreated individuals as the facts of emerging and spreading of these drug-resistant HIV-1 variants. Therefore, our study described the prevalence of surveillance drug-resistant HIV-1 strains at 2.1% among the drug-naïve population in Jiangmen, below the threshold of 5% (the WHO HIVDR Threshold Surveillance protocol). Interestingly, NNRTIs-related DRMs V179D/E occurred the most frequently among the untreated population in our study, which was coincident with the previous research.4, 31, 32 Contrary to V179D/E highly occurring in CRF01_AE, our study showed that V179D/E tended to be found in CRF55_01B strains, implied that DRMs such as V179D/E were shown diversified among the drug- naïve population. Although these mutations merely conferred low or potential drug resistance, they could significantly reduce the replicative fitness to the HIV-1 viruses.33 Our study on TDR surveillance would be essential to prevent the spread of resistant strains for the evaluation of prevention measure and guidance of the first-line antiretroviral regimens. It was rewarding to keep closely tracking and monitoring.
Some limitations should be mentioned in our study. Firstly, it should be noted that the samples collected by convenience did not represent a random sampling. Thus an unknown degree of sampling bias may exist. Secondly, at least two discontinuous genes for each sample were sequenced and analyzed for more complete and detail genetic information if possible in the present study but it would overestimate the actual proportion of recombinant strains due to the reason that some of the previous CRFs were identified as novel URFs. Further analyses of full-length sequences should be performed to reveal the genetic variation of HIV-1 and explore its impact on licensed diagnostic and therapeutic strategy.
ACKNOWLEDGMENTS
The authors would like to thank the Guangdong Province Public Welfare Research and Capacity Building Project (Grant number: 2014B020212007) and the Guangzhou Science and Technology Project (Grant number: 201508020018) for funding this study.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.