Detection of TERT promoter mutation in serum cell-free DNA using wild-type blocking PCR combined with Sanger sequencing in hepatocellular carcinoma
Abstract
Telomerase reverse transcriptase (TERT) promoter mutation is the most frequent genetic alteration in hepatocellular carcinoma (HCC). However, there is currently no suitable highly sensitive method that can detect such mutation using serum cell-free DNA (cfDNA). We analyzed somatic point mutations that substitute cytosine for thymidine at position 228 (C228T), as one of the hotspots of TERT promoter mutations, in serum cfDNA using a highly sensitive detection method of wild-type blocking polymerase chain reaction (WTB-PCR) combined with Sanger sequencing. In TERT promoter mutation sensitivity study, synthetic oligonucleotides were prepared to determine the lowest detection limit of the WTB-PCR, using serial dilutions of mutant-type (MT) DNA in the background of wild-type (WT) DNA. Using this technique, we conducted a longitudinal study in one patient who developed HCC during the follow-up and determined the relationship between HCC and TERT C228T in serum cfDNA. In the sensitivity study, the mutant peak at position 228 was detected at 0.7% or higher but was not detected at 0.6%. Thus, sequencing analysis of WTB-PCR product demonstrated the limit of detection in excess of 0.7% MT DNA in the background of WT DNA. One male patient with HCV-related cirrhosis developed HCC during the follow-up. TERT C228T was negative before the diagnosis of HCC, positive at the diagnosis of HCC and did not increase with advancement of malignancy. We developed a highly sensitive method for detection of TERT promoter mutation using WTB-PCR combined with Sanger sequencing and demonstrated its clinical usefulness in the measurement of TERT C228T in serum cfDNA. Larger studies are needed to confirm these results and establish the clinical utility of this new method.
Research Highlights
-
We developed a highly sensitive method for detection of TERT promoter mutation using WTB-PCR combined with Sanger sequencing and demonstrated its clinical usefulness in the measurement of TERT C228T in serum cfDNA.
Abbreviations
-
- AFP
-
- α-fetoprotein
-
- cfDNA
-
- cell-free DNA
-
- HCC
-
- hepatocellular carcinoma
-
- HCV
-
- hepatitis C virus
-
- LNA
-
- locked nucleic acid
-
- MT
-
- mutant-type
-
- PCR
-
- polymerase chain reaction
-
- TACE
-
- transcatheter arterial chemoembolization
-
- TERT
-
- telomerase reverse transcriptase
-
- WT
-
- wild-type
-
- WTB
-
- wild-type blocking
1 INTRODUCTION
Recent genomic studies have identified telomerase reverse transcriptase (TERT), tumor protein p53, and catenin beta 1 as the most frequently mutated genes in hepatocellular carcinoma (HCC).1-3 TERT promoter mutation is the most frequent genetic alteration in HCC.3, 4 In cirrhosis, TERT promoter mutations were highly related to stepwise hepatocarcinogenesis, with mutations identified in 6% of low-grade dysplastic nodules, 19% of high-grade dysplastic nodules, and 61% of early HCCs.5 Furthermore, since two hotspots of TERT promoter mutations, C228T and C250T, were detected in 94.7% and 5.3% of HCC patients with identified mutations, respectively, a stronger impact for C228T, compared with C250T, is presumed in HCC.6
A recent study reported the detection of TERT promoter mutations in plasma cell-free DNA (cfDNA).7 TERT promoter C228T and C250T mutations were detected by polymerase chain reaction (PCR) using QX200 droplet digital PCR system (Bio-Rad Laboratories, Inc). The prevalence of TERT promoter mutations in The Cancer Genome Atlas HCC specimens was 44.4%. TERT promoter mutations were detected in plasma cfDNAs at similar frequencies in 218 patients with HCC (47.7%), but in only 7 out of 81 (8.6%) patients with cirrhosis without imaging evidence of HCC.7 However, to our knowledge, TERT promoter mutations are still not investigated using serum, not plasma, cfDNA, of patients with HCC.
Sanger sequencing has traditionally been the gold standard in testing for somatic mutations. One of the limitations of Sanger sequencing is its limit of detection (~10%–20% mutant-type [MT] allele in a background of wild-type [WT]).8 This level of sensitivity is inappropriate for detecting low-level somatic mutations that may be present in tissue samples from premalignant tissues or patients with few circulating tumor cells. By replacing conventional PCR with locked nucleic acid (LNA)-mediated wild-type blocking PCR (WTB-PCR) in Sanger sequencing, the sensitivity of up to 0.1% MT in a background of WT can be achieved.9-11 In WTB-PCR, enrichment of MT alleles is achieved through the addition of a blocking oligonucleotide that binds preferentially to WT DNA, thereby preventing the amplification of WT DNA. The MT-enriched WTB-PCR product can then be sequenced. By blocking WT DNA rather than selection for specific mutations, the WTB-PCR allows enrichment of mutations present in minute amounts of cfDNA.12 However, to our knowledge, TERT promoter mutations are still not investigated in patients with HCC using WTB-PCR.
The purpose of the present study was to describe the development of a highly sensitive method for the detection of TERT promoter mutation in serum cfDNA using WTB-PCR combined with Sanger sequencing. We also applied this method clinically to serially quantify changes in TERT promoter mutations in a male patient who developed HCC during the course of follow-up, including local and distant metastasis.
2 METHODS
The study protocol was approved by the Toranomon Hospital institutional review board (#1843). The study was conducted in compliance with the International Conference on Harmonization guidelines for Good Clinical Practice (E6) and the 2013 Declaration of Helsinki.
2.1 Assessment of TERT promoter mutation by WTB-PCR
Blood was withdrawn and the serum sample was frozen at −80°C within 4 hours of the collection then thawed just before analysis. The genome DNA was extracted from 1000 μL of serum with QIAamp Circulating Nucleic Acid Kit (QIAGEN, Tokyo, Japan), and the nucleotide sequences were determined by direct sequencing. The primers used were TERT promoter F (5′-CAGCGCTGCCTGAAACTC-3′; nucleotides 1,295,151-1,295,168 on chromosome 5) and TERT promoter R2 (5′-GGCCGATTCGACCTCTCT-3′; nucleotides 1,295,528-1,295,511 on chromosome 5). The genome sequence of 378 nucleotides was determined. The 228-LNA (5′-GCCCAGCCCCCTCCGGGCCCT-3′; Uppercase letters indicate LNA) was used as the blocking oligonucleotide for TERT promoter at position 228 (TERT228). WTB-PCR master mix was prepared, using 12.5 µL 2X buffer, 5 µL dNTPs, 1 µL forward primer, 1 µL reverse primer, 1 µL blocking oligonucleotide for TERT228, 0.5 µL KOD SYBR qPCR Mix (Toyobo Co, Osaka, Japan), and 3 µL double-distilled H2O to create a final solution volume of 24 µL per reaction. Of this, 1 µL was used for genomic DNA. First denaturation was performed at 94°C for 2 minutes, and 40 cycles of amplification were performed as follows: denaturation for 10 seconds at 98°C, annealing of primers for 30 seconds at 62°C followed by 5 seconds at 72°C, extension for 30 seconds at 68°C, and final extension was performed at 68°C for 7minutes. The PCR-amplified DNA was purified after agarose gel electrophoresis and then used for direct sequencing. The latter was conducted using the dye terminator method. Dideoxynucleotide termination sequencing was performed using the Big Dye Terminator Cycle Sequencing kit (Life Technologies, Tokyo, Japan).
For the TERT promoter mutation sensitivity study, we prepared synthetic oligonucleotides with gBlocks Gene Fragments Libraries (Integrated DNA Technologies, Inc, Tokyo), to determine the lower limit of detection of WTB-PCR. TERT228WT (228C) and TERT228MT (228T) of 748 nucleotides were synthesized (Table S1). Serial dilutions (100%, 90%, 50%, 20%, 10%, 5%, 4%, 3%, 2%, 1%, 0.9%, 0.8%, 0.7%, 0.6%, 0.5%, 0.4%, 0.3%, 0.2%, 0.1%, and 0%) of TERT228MT in background of TERT228WT were performed.
In the present study, we defined TERT C228T “positive” samples as those with mutant peak detected at position 228 (228T), based on the electropherograms in sequencing.
2.2 Validation of WTB-PCR using serum samples from patients with or without HCC
Seven selected HCV patients with HCC in our hospital were cross-sectionally investigated TERT C228T in serum cfDNA, at the time of diagnosis of HCC. Especially, one of them developed HCC during the follow-up and was longitudinally examined for the relationship between HCC and TERT C228T. On the contrary, the other one patient with nonviral chronic liver disease, who did not develop HCC during the follow-up, was longitudinally evaluated TERT C228T, as standard for comparison. Blood tests and imaging studies (ultrasonography or computed tomography) were performed twice annually.
3 RESULTS
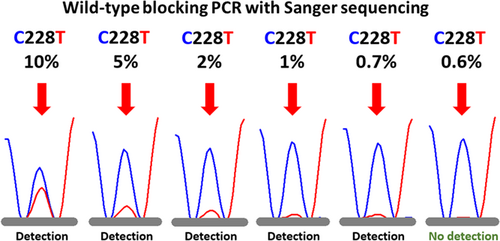
3.1 TERT promoter mutation sensitivity study
Figure 1 shows the electropherograms in the sequencing of the TERT promoter mutation sensitivity study for TERT C228T. The wild peak at position 228 (228C) is shown as the blue peak. The mutant peak at position 228 (228T) (red peak) was detected at 0.7% or higher but not at 0.6% (Figure 1). Thus, the sequencing analysis of the WTB-PCR product demonstrated a detection limit in excess of 0.7% MT DNA in the background of WT DNA.
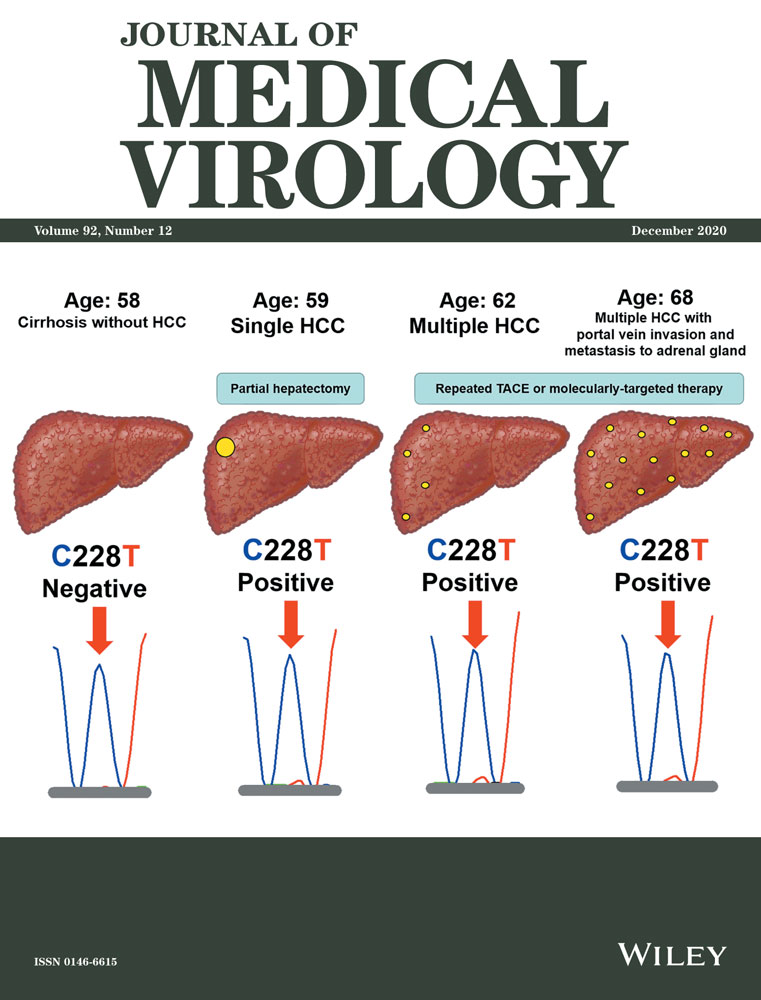
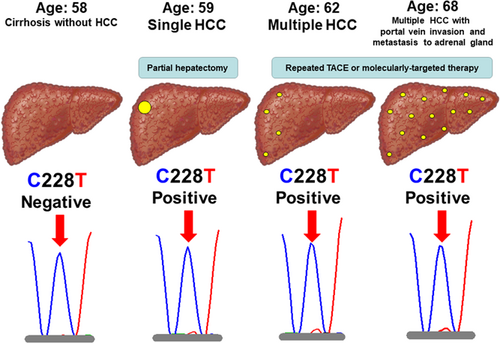
3.2 Clinical validation of TERT C228T in serum cfDNA
In three of seven HCV patients with HCC, TERT C228T in serum cfDNA was positive at the time of diagnosis of HCC. Especially, one male patient with HCV-related cirrhosis developed HCC during the follow-up (Genotype 1b, Viremia levels 5.8 log IU/mL, Core aa70 wild-type, and IL28B rs8099917 genotype TG). He was investigated longitudinally for the relationship between hepatocarcinogenesis and TERT C228T in serum cfDNA. He was diagnosed with cirrhosis but no HCC at 58 years of age, then found to have a solitary HCC a year later, and multiple HCC at 62, followed by portal vein invasion and metastasis to the adrenal gland at 68. TERT C228T was negative before the diagnosis of HCC at 58 years of age, but positive at the time of diagnosis of HCC (Figure 2). The case suggested that TERT C228T can be detected in serum cfDNA of the patient with HCC using WTB-PCR, and the potential utility of WTB-PCR in the clinical setting. Interestingly, the mutant peak (228T) (red peak) did not show any obvious increase following the spread of HCC at 62 (Figure 2). On the contrary, the other one male patient with nonviral and non-cirrhotic chronic liver disease, who did not develop HCC during the follow-up, was longitudinally evaluated, and TERT C228T was negative at 42 and 64 years of age (Figure S1).
4 DISCUSSION
It is still unknown whether TERT promoter mutations can be detected in serum cfDNA of patients with HCC. To our knowledge, the present study is the first to investigate TERT promoter mutations using serum cfDNA rather than plasma cfDNA of patients with HCC. Furthermore, we developed a highly sensitive method for the detection of TERT promoter mutation comprising WTB-PCR and Sanger sequencing. Especially, in the longitudinal study of one patient with HCC, we confirmed the presence of TERT promoter mutation by analyzing serum cfDNA with WTB-PCR, indicating the clinical usefulness of this methodology. Interestingly, the mutant peak (228T) did not increase with the advancement and local and distant spread of HCC in the same patient. This finding suggests that analysis of TERT C228T might help in the early diagnosis of HCC rather than determining the stage of HCC.13 In this regard, a recent study indicated that TERT mutations in plasma cfDNA were associated with poor prognosis in patients with HCC.7 Further longitudinal studies of patients who develop HCC during the follow-up are needed to evaluate the clinical usefulness (eg, early prediction of HCC, survival, and therapeutic reactivity) of TERT C228T in serum cfDNA.
In the present study, seven selected patients with HCC were cross-sectionally investigated TERT C228T in serum cfDNA at the time of diagnosis of HCC. In three of seven patients (42.9%) with HCC, TERT C228T was positive at the time of diagnosis of HCC. The previous report indicated that TERT promoter mutations were detected in plasma cfDNAs at similar frequencies in patients with HCC (47.7%).7 However, we should pay attention to the interpretation of the present results, based on the small number of selected patients with HCC. Further studies of the large number of patients with HCC should be performed to investigate the difference of positive rate of TERT C228T in serum and plasma cfDNA using WTB-PCR.
One limitation of the present study is the lower sensitivity of our method compared with that reported in previous studies using WTB-PCR, with the exception of TERT promoter mutation.9-11 Another limitation is the lack of analysis of TERT C250T mutations. New WTB-PCR-based highly sensitive methods for the detection of TERT promoter mutation, including both TERT C228T and C250T should be developed in the future.
In conclusion, we developed a highly sensitive method for the detection of TERT promoter mutation comprising WTB-PCR and Sanger sequencing. Our preliminary results in a single patient with HCC showed the clinical usefulness of measurement of TERT C228T in serum cfDNA. Further studies of the large number of patients with HCC should be performed to investigate the clinical impact of TERT promoter mutation in serum cfDNA.
ACKNOWLEDGMENTS
The authors thank Rie Mineta and Kenichi Tadokoro for their research assistance. This study was supported in part by a Grant-in-Aid from the Japan Agency for Medical Research and Development (#17fk0210104h0001) (NA), (#18fk0210040h0001) (NA), (#19fk0210058s0901) (NA), and a Grant-in-Aid for scientific research and development from Ministry of Health, Labor and Welfare of Japan (#19HC1001) (NA).
CONFLICT OF INTERESTS
Hiromitsu Kumada has received honoraria from MSD KK, Gilead Sciences, AbbVie Inc, Eisai Co, Ltd, and Dainippon Sumitomo Pharma. Norio Akuta has received honoraria from AbbVie Inc and Gilead Sciences. Yasuji Arase has received an honorarium from AbbVie Inc. Masahiro Kobayashi has received honoraria from Eisai Co, Ltd. All other authors declare no conflict of interests.
AUTHOR CONTRIBUTIONS
NA, FS, MK (Mariko Kobayashi), SF, YK, HS, TH, MK (Masahiro Kobayashi), SS, YA, KI, YS, and HK participated in the design and execution of the study. NA and FS provided the blood samples and analyzed the data. NA wrote the manuscript.
DISCLAIMERS
This paper has not been published or presented at national or international meetings, in part or in its entirety, and is not under consideration by another journal.
ETHICS STATEMENT
The study was conducted in compliance with the International Conference on Harmonization guidelines for Good Clinical Practice (E6) and the 2013 Declaration of Helsinki. The study protocol was approved by the Human Ethics Review Committee at Toranomon Hospital.