Comparison between patients with norovirus-related gastroenteritis and asymptomatic carriers with respect to distribution of antibody-complexed viral particles and intestinal flora
Abstract
Asymptomatic carriers have a major influence on the spreading of norovirus infections. The objective of this study was to examine the characteristics of patients and asymptomatic carriers affected by norovirus-related community gastroenteritis outbreaks. No significant difference between the two groups was observed in terms of the number of norovirus-antibody complexes with respect to total numbers. Principal coordinates analysis of the intestinal flora based on β-diversity analysis, revealed a different bacterial composition between patients and asymptomatic carriers, particularly regarding the genera Pseudomonas, Bacteroides, and Erwinia, as well as the Ruminococcaceae family. Although the proportional changes between these intestinal microorganisms were not sufficient to explain gastroenteritis symptoms, they represent possible markers shared by asymptomatic norovirus carriers.
1 INTRODUCTION
The Norovirus genus, which is part of the Caliciviridae family, comprises single-stranded, positive-sense RNA viruses. norovirus is a major causative agent of viral gastroenteritis in both community and sporadic cases, and a major cause of nosocomial outbreaks. Asymptomatic carriers play an important role in the spreading of norovirus in community gastroenteritis outbreaks. It is believed that there is a relationship between food handlers, who are asymptomatic carriers, and community gastroenteritis outbreaks.1 Based on the Tokyo Metropolitan Government Annual food poisoning summary, sources of food-borne community gastroenteritis outbreaks in Tokyo between 2003 and 2015, were traced mainly to food handlers who had gastroenteritis symptoms or were suspected to be asymptomatic carriers (Table 1). Asymptomatic carriers are thought to contaminate the equipment and facilitate indirect person-to-person transmission. Quantitative analyses comparing fecal specimens of patients and asymptomatic carriers from norovirus-related food-borne and nosocomial gastroenteritis outbreaks have been performed and reported previously.2, 3 They suggested no significant differences in viral copy numbers in fecal specimens between the two groups. However, asymptomatic carriers in those studies could not be adequately characterized owing to a lack of collected specimens. The aim of the present study was to compare the characteristics of patients and asymptomatic carriers affected by norovirus-related viral gastroenteritis community outbreaks. To verify the influence of acquired IgA antibodies and the intestinal flora, virus-antibody complexes were examined in fecal specimens and 16S metagenomics analysis was performed.
| 2000 | 2001 | 2002 | 2003 | 2004 | 2005 | 2006 | 2007 | 2008 | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | 2015 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Shellfisha | 14 | 11 | 23 | 15 | 9 | 12 | 7 | 2 | 7 | 11 | 26 | 21 | 15 | 2 | 6 | 17 |
| Food handlerb | 5 | 3 | 6 | 17 | 15 | 19 | 36 | 23 | 24 | 26 | 42 | 28 | 42 | 23 | 15 | 39 |
| Unknown | 2 | 3 | 1 | 2 | 2 | 1 | 2 | 2 | 2 |
- Data are based on the Tokyo Metropolitan Government Annual food poisoning summary.
- a Outbreaks which had history of consumption of eating of shellfish.
- b Outbreaks which were suspected to relate with food handler.
2 MATERIALS AND METHODS
2.1 Samples
Stool specimens were collected from 33 patients and 23 asymptomatic carriers in nine norovirus-related food-borne community gastroenteritis outbreaks that occurred in Tokyo between April 2012 and April 2013. All outbreaks were observed in adults. Each specimen was collected along with data for patients or asymptomatic carriers identified them as either adults or infants. In addition, 20 specimens belonging to five asymptomatic carriers in whom norovirus was detected, were collected 7, 10, 13, 36, and 45 days after consumption of the contaminated food. These served to confirm negativity to norovirus in Tokyo between February 2010 and February 2011. All specimens were used for nucleic extraction, and subsequent comparison of a region of the virus-antibody complex, and 16S metagenomics analysis. This study was evaluated and approved by the Ethics Review Committee of Tokyo Metropolitan Institute of Public Health.
2.2 Nucleic acid extraction and reverse transcription
Freshly prepared 10% fecal suspensions in phosphate-buffered saline (PBS) were centrifuged at 1,300g for 5 min at 4°C. Nucleic acids were extracted from 140 μL of each supernatant using the QIAamp Viral RNA mini kit (QIAGEN, Hilden, Germany) following the manufacturer's instructions, to evaluate the total copy numbers of norovirus in fecal specimens and perform 16S metagenomics analysis. To evaluate antibody-virus complexes, supernatant from 10% suspensions of fecal specimens pre-incubated with anti-IgA (secretory) antibodies (MBL, Nagoya, Japan) was diluted with binding buffer, applied to an IgG-specific binding column (Albumin & IgG Depletion SpinTrap; GE Healthcare, Little Chalfont, UK), and incubated at 25°C for 5 min. The column was centrifuged, rinsed with binding buffer, and incubated after addition of buffer AVL (QIAamp Viral RNA mini kit). Buffer AVL was collected in a microtube by centrifugation at 1,400g for 30 s at 25°C. The eluted buffer should include nucleic acids from viral particles complexed with IgA antibodies. Next, nucleic acids were extracted according to the manufacturer's instructions of the QIAamp Viral RNA mini kit and stored at −80°C until use. The reverse transcription reaction used 12 μL of the extracted nucleic acid with 18 μL of a mixture containing 0.5 mM deoxynucleotide triphosphate (dNTPs), 5 mM dithiothreitol (DTT), 16 U RNase inhibitor (Nacalai Tesque, Kyoto, Japan), 1 μg of random hexamers, 300 U of SuperScript II reverse transcriptase (Thermo Fisher Scientific, Waltham, MA), and reaction buffer. The reaction mix was then incubated at 42°C for 1 h.
2.3 Real-time polymerase chain reaction (PCR)
Norovirus was detected by real-time PCR4 using the COG1F/1R primer pair and RING1TPa probe for norovirus GI, as well as the COG2F/2R primer pair and RING2TP probe for norovirus GII. After reverse transcription, real-time PCR was performed using 5 µL of cDNA and 20 µL of a reaction mixture containing TaqMan Universal Master Mix (Thermo Fisher Scientific), 1 µM of each primer pair, and 0.15 µM of each probe. Real-time PCR was carried out using an ABI PRISM 7900HT sequence detection system (Thermo Fisher Scientific), with the following reaction conditions: a 10 min denaturation step at 96°C, followed by 45 cycles at 96°C for 15 s and 56°C for 1 min. Quantitative analysis was performed using standard DNA manufactured by the National Institute of Infectious Diseases (Tokyo, Japan). Next, the number of viral copies present in norovirus patients and asymptomatic carriers was determined. Additionally, norovirus from each detected case was amplified using G1SKR/COG1F and G2SKR/COG2F primer pairs.5 Sequence identities were determined using the norovirus typing tool (RIVM: http://www.rivm.nl/en/Topics/N/NoroNet).
2.4 16S metagenomics analysis
To compare the intestinal flora in norovirus patients and asymptomatic carriers, 16S metagenomics analysis was performed according to the 16S Metagenomic Sequencing Library Preparation guide (https://jp.illumina.com/areas-of-interest/microbiology/human-microbiome-analysis.html). Extracted nucleic acids were amplified with 16S amplicon PCR primers located within the V3 and V4 region. Thirty-seven of the 16S amplicon PCR positive cases (17 patients and 20 asymptomatic carriers belonging to seven norovirus-related outbreaks) were purified and analyzed by MiSeq (Illumina, San Diego, CA). Collected sequences were compared using a MiSeq reporter (Illumina), and β-diversity analysis was performed to indicate the ratio between regional and local diversity using QIIME software.6 The unweighted pair group method with arithmetic mean (UPGMA) tree was constructed based on β-diversity analysis. Additionally, principal coordinates analysis (PCoA) was performed on the β-diversity data. The order defining the proportion of variance of principal components was: PC1, PC2, and PC3.
3 RESULTS
3.1 Comparison of norovirus-IgA complexes in fecal specimens
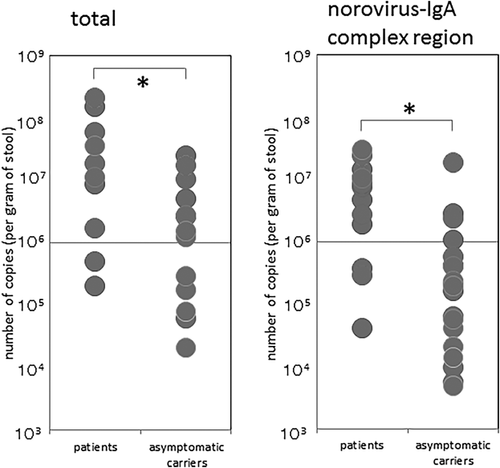
All outbreaks were classified as GII (one GII.2, five GII.4, and three GII.13). The medians for total norovirus copy number (per gram of stool) in patients and asymptomatic carriers were 17436480 and 1259238.2, respectively. Significant difference was detected between patients and asymptomatic carriers using the non-parametric Mann-Whitney u test (P < 0.01). Norovirus was detected by real-time PCR in the norovirus-IgA complex region in all specimens. This region was captured by an IgG-specific biding column which was pre-incubated with anti-IgA antibody. The specimens were collected at the earliest 3 days after consumption of the suspected food. The medians of copy number (per gram of stool) for the norovirus-IgA complex region in patients and asymptomatic carriers was 7553341.7 and 196428.4, respectively. Significant difference was detected using the non-parametric Mann-Whitney u test (P < 0.01) (Figure 1). However, no significant difference between the two groups was observed in terms of the numbers of norovirus-IgA complexes with respect to total norovirus numbers using the non-parametric Mann-Whitney u test (P = 0.540). The medians for norovirus-IgA complex region/total norovirus copy number in patients and asymptomatic carriers were 21.9% and 19.3%, respectively. Moreover, no difference was observed on the basis of genotype either. norovirus copy numbers were inversely proportional to the number of norovirus-antibody complexes (data not shown).
3.2 Comparison of intestinal flora
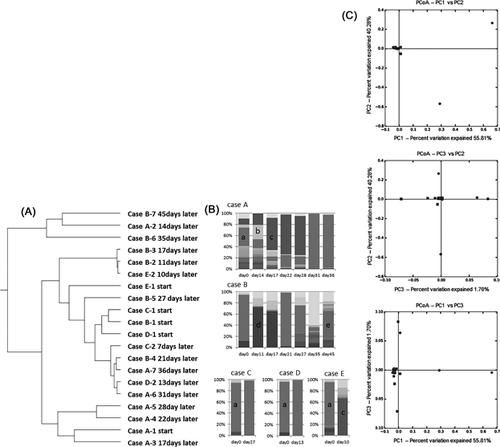
The UPGMA tree was calculated on the basis of results from the β-diversity analysis in asymptomatic carriers who were still infectious with norovirus from ten to 45 days after the first norovirus was detected, but was not branched independently for each patient. Instead, the UPGMA tree was constructed in such way that it enabled the observation of the distribution over time. Alterations in the composition of the intestinal flora were observed in three out of five cases post infection (Figure 2B). Pseudomonas was the dominant genus at the start of each time course. After 14 days, Pseudomonas was replaced by the genera Ruminococcus and then Stenotrophomonas (at 17, 22, and 28 days) (Case A), by the genera Agrobacterium (at 11 and 17 days) and Faecalibacterium (at 45 days) (Case B), or by the genus Agrobacterium (at 10 days) (case E) (Figure 2B). Using results from the β-diversity analysis between patients and asymptomatic carriers, PCoA revealed the clustering of distributions based on PC1 versus PC2, PC1 versus PC3, and PC2 versus PC3 (Figure 2C). No differences were found on the basis of norovirus genotypes. However, significant differences were observed between patients and asymptomatic carriers by g-test for the genera Pseudomonas (P < 0.01), Bacteroides (P < 0.01), Erwinia (P < 0.01), Agrobacterium (P = 0.024), the Ruminococcaceae family (P < 0.01), and the RF32 order (P = 0.031). Specifically, the Pseudomonas genus was dominant in norovirus patients; whereas the Bacteroides, Erwinia, and Agrobacterium genera, Ruminococcaceae family, and RF32 order were dominant in asymptomatic carriers.
4 DISCUSSION
Asymptomatic norovirus carriers exert a major influence on the transmission of norovirus in community gastroenteritis outbreaks and in the contamination of equipment and facilities. While epidemic studies of asymptomatic norovirus infections have been reported previously,7-9 the mechanism of asymptomatic infection has remained unknown.
Here, real-time PCR was employed for high-specificity detection of norovirus from clinical specimens. No significant difference was observed between patients and asymptomatic carriers with the respect to the rate of norovirus-IgA complex region/total number of norovirus using the non-parametric Mann-Whitney u test, and in terms of norovirus copy numbers by using t-test, which is consistent with previous reports.2, 3 Nevertheless, it should be noted that aggregated or assembled virus particles in fecal specimens, which seemed to bind to antibodies, had sometimes been observed by electron microscopy examination.10 Indeed, acquired norovirus-specific antibodies in serum and IgA antibodies in breast milk were previously reported,11, 12 but information on fecal IgAs was limited.13 Acquired IgA antibodies are thought to act by providing protection from infection or reducing symptoms. However, in this study, no significant difference was observed in the rate of norovirus-IgA complexes between patients and asymptomatic carriers. Moreover, the specimen in which the earliest norovirus-antibody complex was detected, was collected after 3 days after consumption of the contaminated food. A period of 3 days following consumption of norovirus contaminated food was likely insufficient to allow binding to infected norovirus particles and thus construct antibodies specific to the virus. The detected region on the norovirus-antibody complex can potentially include both the norovirus-IgG complex and the norovirus-IgA complex in fecal specimens. Accordingly, the antibodies observed to bind to norovirus particles might have developed by cross-reaction or non-specific reaction of acquired IgA antibodies over the time course of the experiment. Here, norovirus copy numbers were inversely proportional to the number of norovirus-antibody complexes (data not shown). Acquired antibodies are thought to result in a reduction of norovirus titers. In long-term norovirus shedding cases, evaluation of the norovirus-antibody complex was inadequate due to the low number of norovirus copies at each time point.
The influence of the intestinal flora on norovirus infection was previously reported.14-18 However, a comparison with asymptomatic carriers had not been performed. In previous reports,14-18 commensal bacteria were found to enhance enteric virus infection.15 Histo-blood group antigen (HBGA)-expressing bacteria were suspected to protect norovirus infections owing to their binding ability.16 Consumption of probiotic fermented milk was found to diminish fever symptoms for norovirus-related gastroenteritis,17 which may not come as a surprise given that the gut microbiome profile is associated with development of diarrheal symptoms.18 Accordingly, the intestinal flora is likely related to gastroenteritis symptoms17 and differs based on the pathogen involved.18 In this study, patients with long-term norovirus infection but lacking any symptoms were examined; replacement of Pseudomonas with Stenotrophomonas was observed in two cases (cases A and E) (Figure 2B). Stenotrophomonas were previously reported as HBGA-positive bacteria.19 This result supports the role of the intestinal flora in the reduction of viral titers (Figures 2A and 2B). The same substitution of the bacterial flora was observed in two out of five cases (Figure 2B); however, loss of dominance by the Pseudomonas genus occurred also in the other three cases. These specimens were collected mostly three to 5 days after consumption of contaminated food; the position in the time course seemed to be similar to that of cases with long-term norovirus infection. Clustering of the distribution based on each patient and asymptomatic carrier was observed by PCoA analysis based on β-diversity data (Figure 2C). Even though this investigation was performed with specimens collected 3days or later after the outbreak ‘s onset, a significant difference was observed in some of the fauna between patients and asymptomatic carriers. This result was based on analyzing the same points in the time course after consumption of contaminated foods. Any association between the detected intestinal flora and significant differences in patient symptoms had not been adequately addressed in previous studies of norovirus-related gastroenteritis.14-18 The characteristics of asymptomatic carriers have long been unclear owing to a deficiency in specimen collection. In contrast, the present findings suggest that asymptomatic carriers with norovirus infection share common characteristics.
Further studies, such as those examining the characteristics of virus-binding antibodies, differing populations of detected intestinal flora, and host markers of infection, might help evaluate how an infected person can become an asymptomatic carrier.